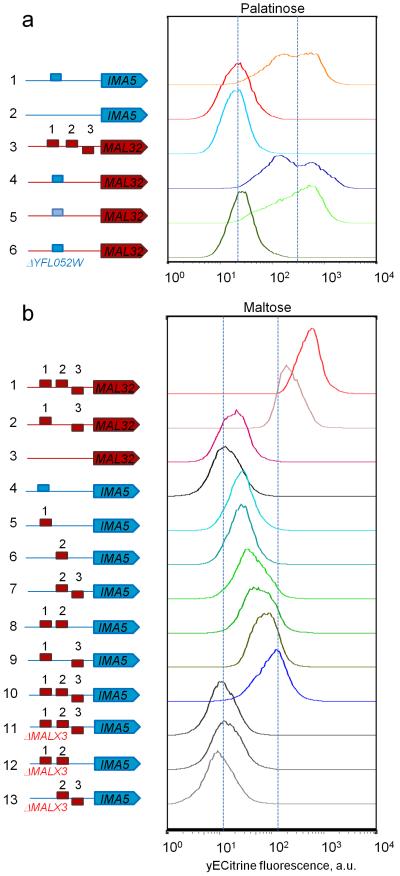
Figure 3. Maltose and palatinose-specific MalR regulators have different DNA binding sites.
Representative flow cytometry histograms of populations of fluorescent reporter strains carrying different types of DNA binding sites in the promoters of a maltose-specific gene MAL32 (red block arrow), or a palatinose-specific IMA5 (blue block arrow), grown in maltose or palatinose. CGG-containing binding sites found in promoters of palatinose-specific genes are depicted as blue rectangles, CGC-containing DNA binding sites found in promoters of maltose-specific gene are depicted as red rectangles. (a) DNA-binding specificity of the palatinose-specific regulator Yfl052w. (1) A fluorescently-tagged IMA5 gene under its native promoter shows a normal level of IMA5 expression activated by Yfl052w in palatinose. (2) Deletion of the Yfl052w binding site abolishes the expression of IMA5. (3) A fluorescently-tagged maltose-specific MAL32 under its native promoter shows no expression on palatinose and is used to quantify autofluorescence. Introduction of a Yfl052w binding site from the promoter of IMA5 (4) or IMA1 (5) in front of the fluorescently-tagged MAL32 gene makes the expression of this gene responsive to palatinose. (6) Deletion of YFL052W abolishes the expression of the MAL32 gene from (4). (b) DNA-binding specificity of the maltose-specific regulator Malx3. (1) A fluorescently-tagged MAL32 under its native promoter shows a normal level of MAL32 expression activated by Malx3 in maltose. (2) Deletion of one Malx3 binding site decreases the expression levels of fluorescently-labeled MAL32. (3) Deletion of all three Malx3 binding sites abolishes the expression of MAL32. (4) A fluorescently tagged IMA5 gene under its native promoter is used to show autofluorescence. Introduction of one (5),(6), two (7)-(9) or three (10) Malx3 binding sites in the promoter region of a fluorescently-tagged IMA5 gene makes the expression of this gene responsive to maltose, with increasing number of binding sites resulting in higher expression levels. (11)-(13) Deletion of the maltose-specific regulator MALX3 abolishes the expression of strains from (7),(8),(10). Each experiment was repeated at least three times with two biological replicates.