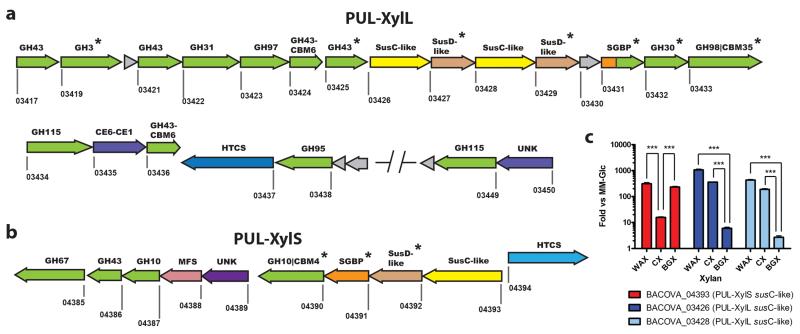
Figure 2. Schematic of the B. ovatus xylan PULs and differential expression during growth on different xylans.
a,b schematic of PUL-XylL and PUL-XylS, respectively. Each gene is drawn to scale as a rectangle with its orientation indicated by the arrow head. The numbers below each gene are its locus tag (BACOVA_XXXXX). Genes encoding known or predicted functionalities are colour-coded and, where appropriate, are also annotated according to their CAZy family number: glycoside hydrolase (GH; green), carbohydrate esterase (CE; purple), carbohydrate-binding module (CBM). Surface located proteins are marked with an asterisk. SGBP = surface glycan binding protein are coloured orange, or, if also in a CAZy family (BACOVA_03431; inactive GH10), are coloured half orange, half green. UNK = unknown (purple), but distant similarity to CE6 carbohydrate esterases. HTCS = hybrid two component system (light or dark blue). MFS = transporter of the major facilitator superfamily (pink). Grey = unknown function (note, there is a structure of BACOVA_03430, PDB accession code 3N91). SusC-like (yellow) and SusD-like (light tan) proteins are a defining feature of PULs and are responsible for import of complex glycans across the outer membrane18. SusC-like proteins are TonB-dependent porins, while SusD-like proteins are surface lipoproteins that likely function to deliver the target glycan to their partner SusC. c, Cells were grown on minimal media with polysaccharide as the sole carbon source, and levels of different susC transcripts (locus tags shown; used as a proxy for expression of the whole PUL) from each PUL were quantified by qRT-PCR. The y-axis shows the Log fold-change relative to a minimal media-glucose reference; x-axis labels indicates the xylans used. WAX – wheat arabinoxylan, CX – corn xylan, BGX – birchwood glucuronoxylan. CX is a highly complex xylan compared to WAX and BGX. All data were analysed by one-way ANOVA followed by Tukey’s Multiple Comparison Test (*** = P≤0.001).