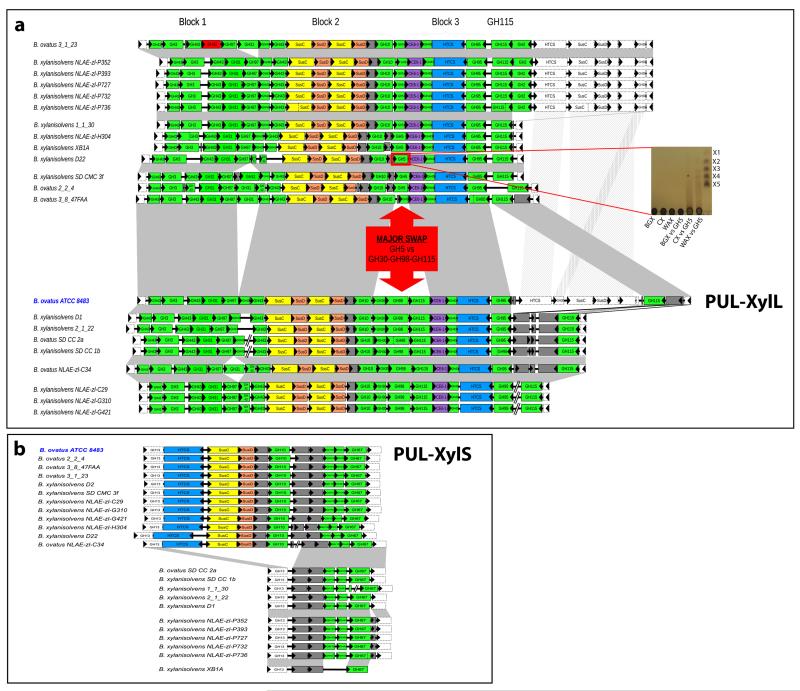
Figure 8. Conservation of the B. ovatus ATCC 8483 xylan PULs in other Bacteroides species.
CAZymes (green, CBM modules were omitted for clarity), susC-like (yellow), susD-like (orange) and the HTCS sensor-regulator (blue) genes are colour-coded. Genes encoding proteins of unknown function or non-CAZymes (e.g. surface glycan binding protein and MFS carbohydrate importer in PUL-XylS) are shown in grey. a, PUL-XylL. Vertical dotted lines indicate split gene models, while missing gene models are shown in red. Microsyntenic segments are depicted by grey connectors, and include adjacent conserved non-PUL members (shown with dotted lines). Eighteen genes are conserved across all strains, and 17 of these form three highly conserved blocks indicated on top. The major difference between the two groups of the GH98 for GH5_21 swap is indicated. Inset shows a TLC of the activity of the recombinant B. xylanisolvens XBA1 GH5_21 enzyme (BXY_29320; boxed in red) against different xylans (1μM final enzyme vs 0.5% xylan in 20 mM sodium phosphate buffer, pH 7.0, 16 h at 37 °C). The B. xylanisolvens GH5 enzyme displays endo-xylanase activity against CX, producing a smear of large oligosaccharide products similar to the product profile observed for the B. ovatus GH98 enzyme, BACOVA_03428. The GH5 xylanase also hydrolyses WAX, unlike the GH98 enzyme, but displays no activity against BGX. X1-X5 indicates xylooligosaccharide standards. b, PUL-XylS. The lower part of the panel shows the strains that contain only the second half of PUL-XylS that encodes the intracellular (periplasmic and cytoplasmic) enzymes, MFS class carbohydrate importer and HTCS.