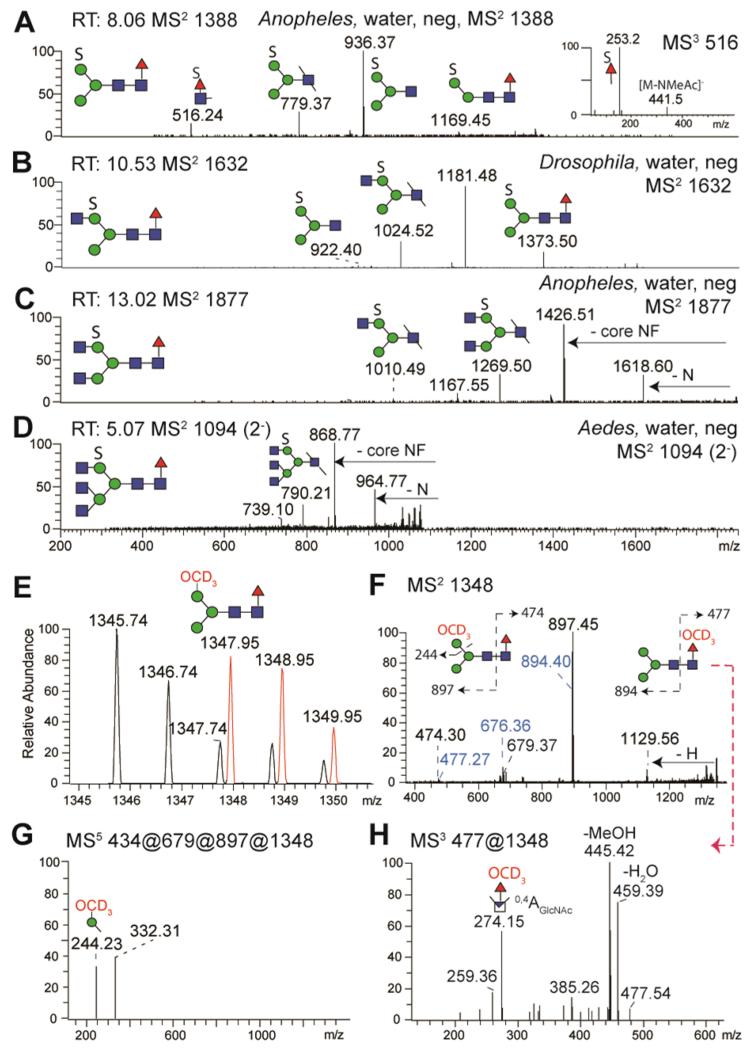
Figure 8. Detection of sulphated N-glycans by NSI-MS.
PNGase F released sulphated N-glycans from Anopheles, Aedes and Drosophila were enriched upon permethylation in the water-phase and analysed by NSI-MS; automated TIM MS2 negative ion mode spectra at certain scan run times (RT) are shown for the major sulphated N-glycans (A-D). After solvolysis and repermethylation with deuterated iodomethane CD3I, a new deuterated glycan isotopic distribution (in red, E) was detected in positive ion mode of the organic phase which is 2.2 a.m.u. different from the neutral N-glycan (H3N2F, m/z 1345). The positive ion mode MS2 spectrum (F) shows daughter ions indicating the loss of hexose (m/z 1129) and monofucosylated core N-acetylglucosamine (m/z 897). MS5 indicates that the deuteromethyl group is associated with hexose (m/z 244; G). The m/z 516 ion in panel A is consistent with the presence of sulphated fucose on a small proportion of the core fucosylated glycans (see MS3 inset and also Supplementary Figure 6). The MS2 of the m/z 1348 repermethylated species further indicates the presence of low abundance fragments (F, in blue) originating from an originally sulphated core fucose; (H) MS3 of the m/z 477 ion results in an m/z 274 fragment indicative of a perdeuteromethylated fucose.