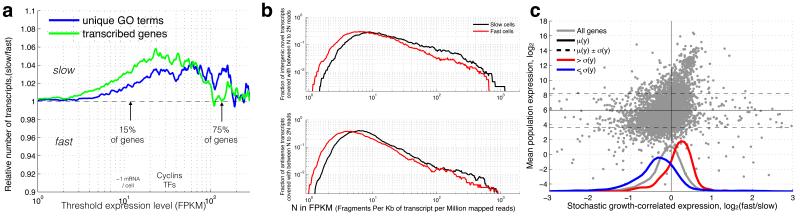
Figure 2. More transcriptional diversity in slow growing subpopulations.
(a) At low expression (< 5 FPKM (Fragments Per Kilobase of transcript per Million mapped reads)), slow and fast growing cells express similar numbers of transcripts, but at medium (5-30 FPKM), slow growing cells express both more genes and more unique gene functions (paired ttest p<1e-35 for transcripts & GO terms). (b) The slow growing subpopulation expresses more unannotated transcripts (paired ks-test p=5.36*10−10) and antisense transcripts (paired ks-test p=1.34*10−22) at >10FPKM. (c) Highly expressed genes (higher than one standard deviation, red) are up-regulated (paired ks-test p=1.1*10−63), while lowly expressed genes (lower than one standard deviation, blue) tend to be down-regulated with increasing subpopulation growth rate (paired ks-test p=4.6*10−35). Y-axis shows the average expression level in all measured populations. The X-axis shows expression change from slow to fast subpopulation growth, computed as the log2 ratio.