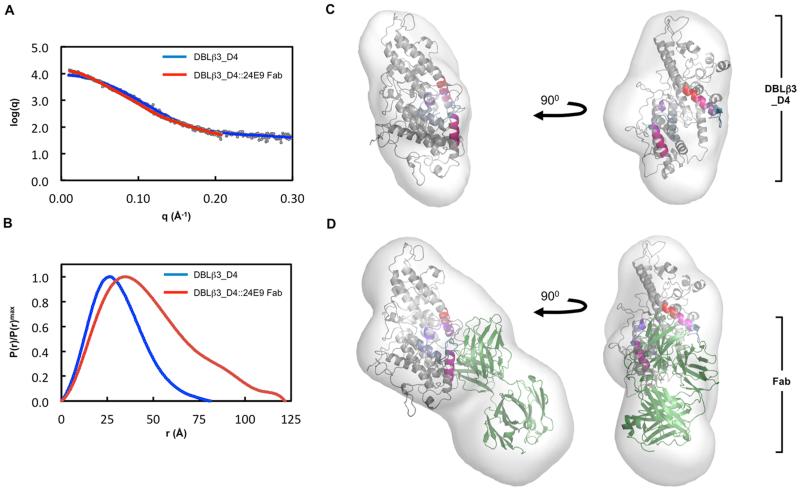
Fig. 8. Molecular architecture of the PFD1235w DBLβ3_D4:24E9 Fab complex.
(A) Theoretical scattering curves derived from ab initio models (solid lines) of DBLβ3_D4 and DBLβ3_D4::24E9 Fab superimposed on the experimental scattering data (circles). (B) Normalized distance distribution function P(r) for DBLβ3_D4 and DBLβ3_D4::24E9 Fab. (C) and (D) The homology model of DBLβ3_D4 was docked into ab initio envelopes of (C) DBLβ3_D4 alone and (D) the DBLβ3_D4::24E9 Fab complex. The peptides identified as being protected in the DBLβ3_D4::24E9 Fab complex are color coded as in Fig. 5. The structure of a mouse Fab fragment (green, PDB ID 3GK8) was used as a model for 24E9 Fab.