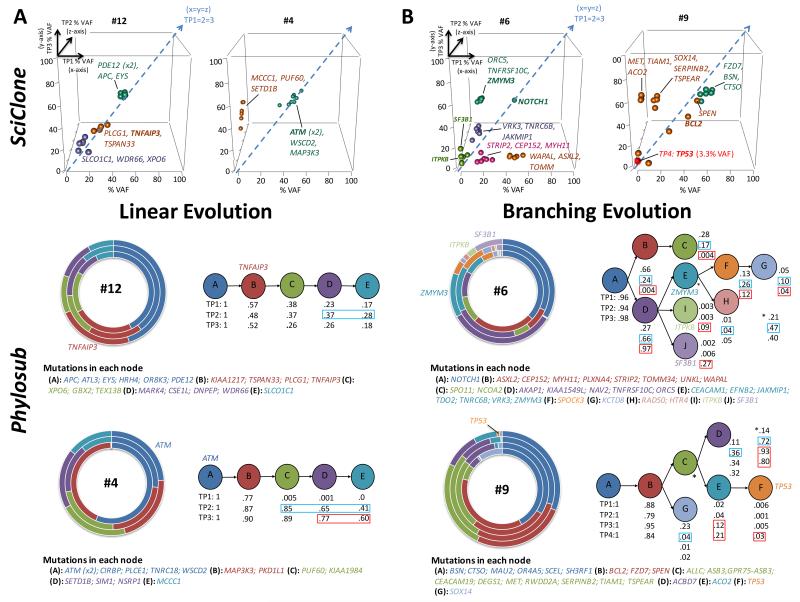
Figure 3. PhyloSub analysis of TDR results in predicted linear and branching clonal evolutionary pattern.
By columns (A) Two patient examples (pt-12 and 4) of linear evolution path, (B) Two patient examples (pt-6 and 9) of complex branching trajectories. (A) & (B) top panel: XYZ scatter-graphs displaying the SciClone mutation clustering analysis on TDR datasets from sequential tumour time points TP1 (x-axis; first tumour sample), 2 (z-axis; progression) and 3 (y-axis; post-treatment). Data point symbols denote a distinct mutation cluster and the x=y=z line is displayed as a dashed blue arrow and denotes no change in the tumor purity-adjusted %VAF of mutation clusters between time points (clonal equilibrium). Selected gene symbols are displayed adjacent to its corresponding mutation cluster. (A) & (B) bottom panel: Concentric pie charts (each layer, inner to outer, is an early to later sampling time point) displaying imputed SNV population frequencies at each phylogenetic node. Predicted phylogenetic tree structure (best model shown), with population frequencies for each node from Phylo-sub analysis. Blue and red boxes denote large changes SNV population frequencies prior to and after first-line treatment, suggesting ongoing clonal dynamics and selection by therapy, respectively. Selected gene symbols are displayed adjacent to the corresponding segment of the pie chart or phylogenetic node.