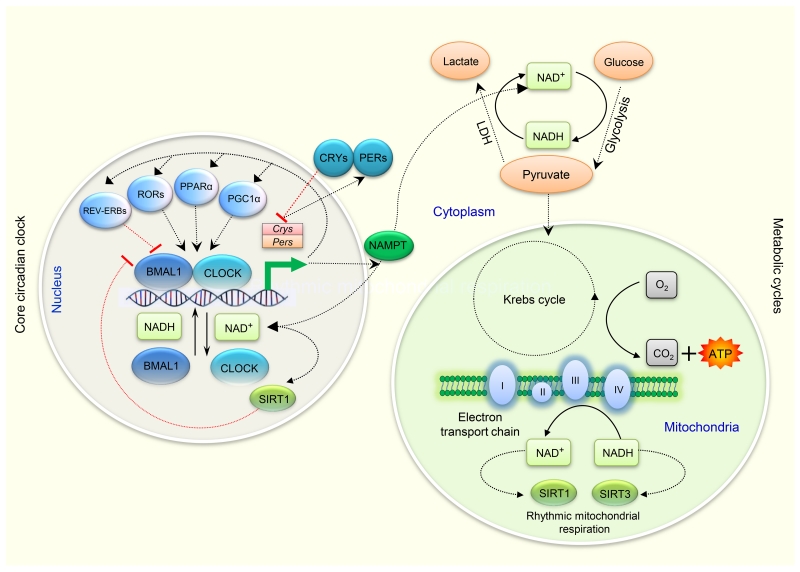
Figure 3. Cross-talk between the circadian and metabolic clocks.
Coupling mechanisms between the circadian and metabolic oscillators are miscellaneous. There are a series of transcription/translation feedback loops in the core clock mechanism. CRY proteins (along with the PER proteins) function as the negative regulators for maintenance of circadian rhythms. PPARα and PGC-1α stimulates expressions of clock genes, while RORs regulate Bmal1 transcription through formation of a feedback loop involving RORα and REV-ERBα. Core clock proteins such as BMAL1 and CLOCK (NPAS2 substitutes for CLOCK in some brain regions (not shown)) regulate the rate limiting steps of NAD+ biosynthesis [58, 62], while the DNA binding affinity of BMAL1 and CLOCK is controlled by the intracellular NAD+/NADH ratio [45]. LDH plays a crucial role in increasing the cellular concentration of NAD+. NAD+-dependent deacetylases e.g. SIRT1 or SIRT3 regulate circadian clock gene expression [59, 64]. NAMPT acts as a rate-limiting enzyme in mammalian NAD+ biosynthesis and its expression is also regulated by the core clock genes [62]. (Details for these possible connecting components between the circadian oscillators and various metabolic processes have been summarised in Table 1). Abbreviations: BMAL1, Brain and muscle ARNT-Like 1; CLOCK, circadian locomotor output cycles kaput; Cry, Cryptochrome; LDH, Lactate dehydrogenase; NAD, Nicotinamide adenine dinucleotide; NAMPT, Nicotinamide phosphoribosyl-transferase; NPAS2, Neuronal PAS domain protein 2; Per, Period; PPAR, Peroxisome proliferators–activated receptor; PGC-1α, PPAR gamma coactivator-1 alpha; ROR, Retinoic acid orphan receptors; SIRT 1, Sirtuin 1; SIRT 3, Sirtuin 3.