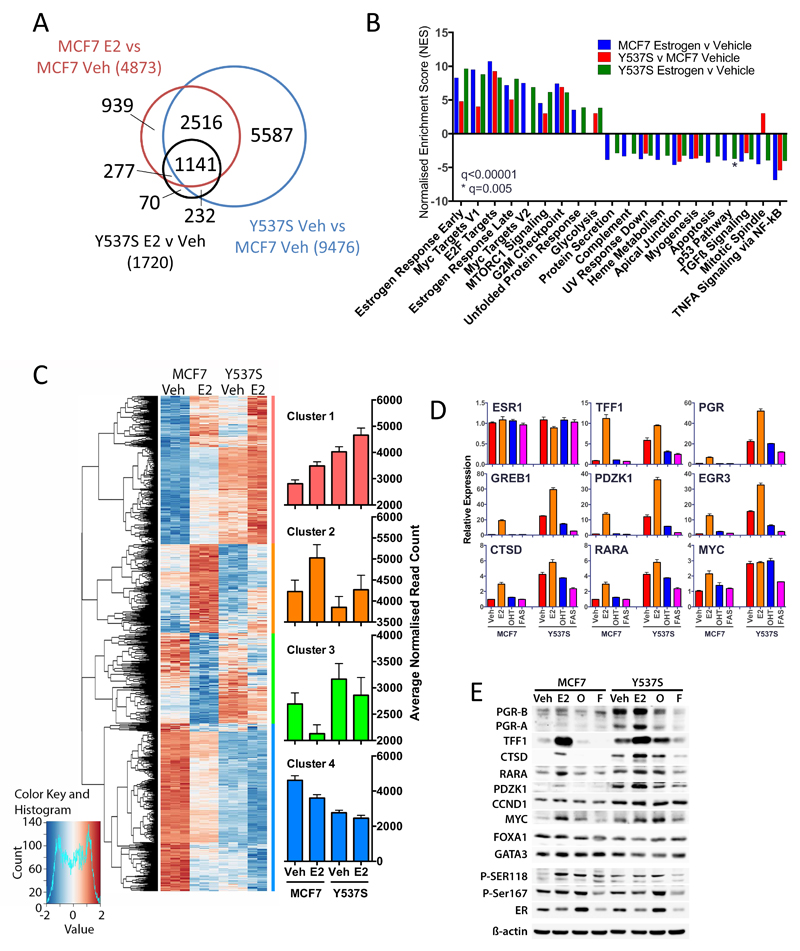
Figure 4. The Y537S mutation promotes estrogen-independent expression of ER target genes in breast cancer cells.
(A) RNA-seq was performed using three replicate samples of hormone-depleted MCF7 and MCF7-Y537S cells, following addition of vehicle or estrogen for 8 hours. Shown is a Venn diagram comparing differentially regulated genes (padj<0.05) identified from RNA-seq data. (B) The bar chart shows the normalised enrichment scores (NES) from Gene set enrichment analysis (GSEA) for “Hallmark” signaling pathways that are significantly up- or down-regulated (q<0.00001) in pair-wise comparisons of the differentially regulated gene sets. (C) Hierarchical cluster analysis of genes that are differentially regulated in MCF7 cells ±E2 (padj<0.05) and for which an ER binding site is observed within 100kb of the transcription start site. Barcharts show average normalised counts for all genes in the clusters. (D) E2 (1 nM), OHT (10 nM) or FAS (10 nM) were added to hormone-depleted MCF7 and MCF7-Y537S cells. Shown is RT-qPCR analysis of gene expression using RNA prepared 16 hours following addition of ligands, relative to the vehicle treated MCF7 cells (n=3). (E) Immunoblotting was performed using cell lysates prepared 24 hours following addition of ligands.