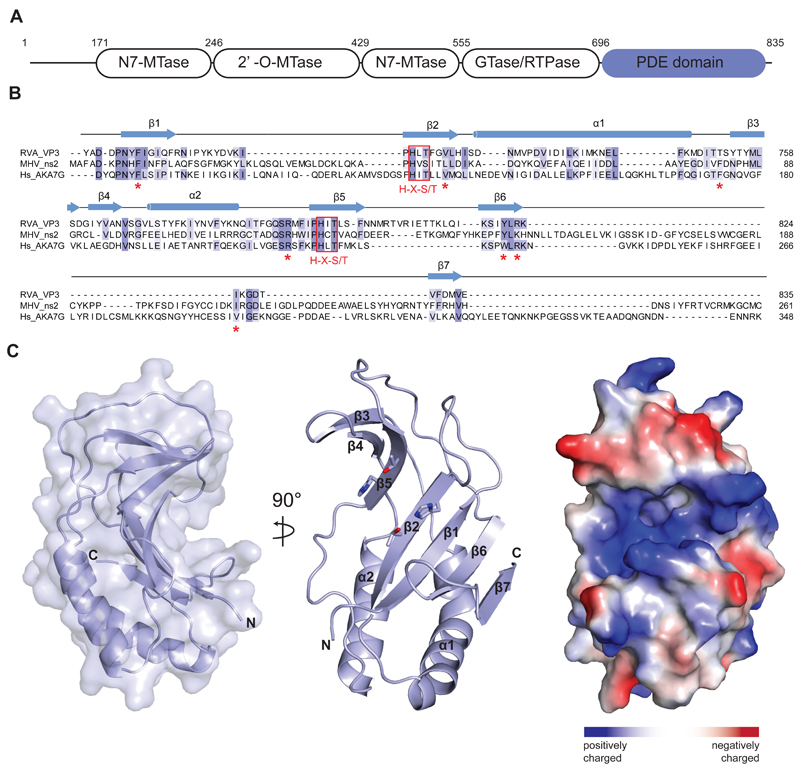
Figure 1.
(A) Domain architecture of group A rotavirus VP3 protein (UniProt ID A2T3S1). The protein consists of an N7-methyltransferase (N7-MTase), 2’-O-methyltransferase (2’-O-MTase) and guanylyltransferase/RNA-triphosphatase (GTase/RTPase) domains, followed by a C-terminal phosphodiesterase (PDE) domain spanning residues 696–835. (B) Multiple sequence alignment for 2H phosphoesterase superfamily members displaying 2’,5’-phosphodiesterase activity. Primary sequences of RVA VP3 (Uniprot ID A2T3S1), MHV ns2 (UniProt ID P19738) and human AKAP7 (Hs_AKA7G, UniProt ID Q9P0M2) proteins were aligned using MAFFT34 with the E-INS-I strategy. Conserved catalytic His-X-Ser/Thr motifs (red boxes) and active site residues (red asterisks) are highlighted. Secondary structure elements of RVA VP3 are indicated above the sequence. (C) Crystal structure of the VP3 PDE domain. Left and middle: Two perpendicular views shown in cartoon representation. Histidine and threonine sidechains of the two His-X-Thr catalytic motifs are shown as sticks. Right: Surface representation colored according to electrostatic surface potential.