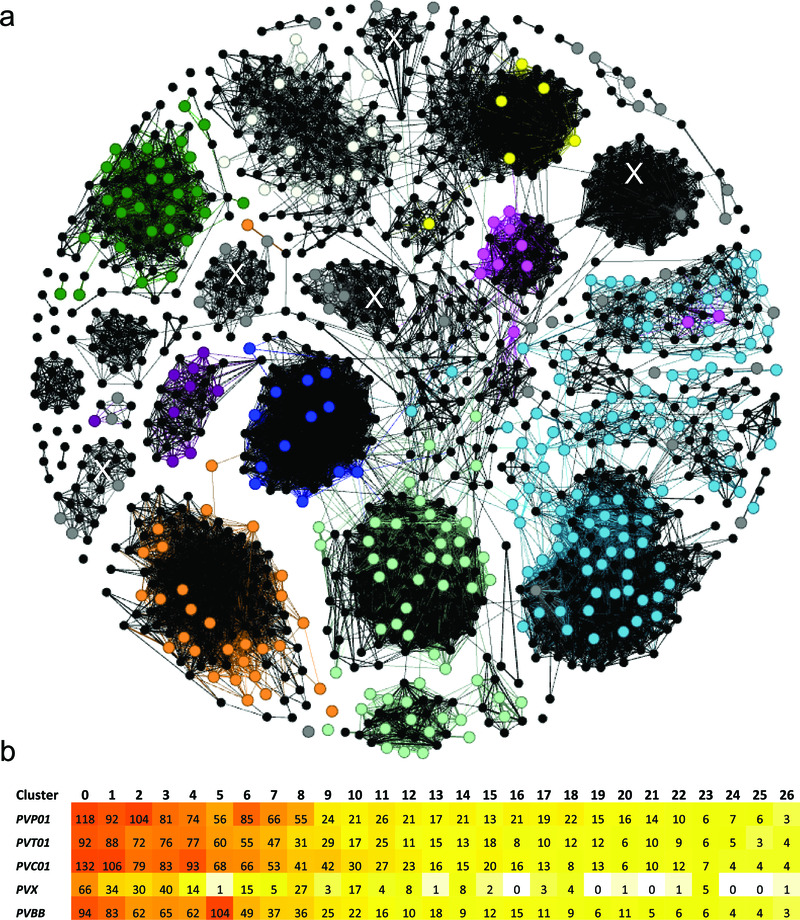
Figure 2. Cluster analysis illustrating the relatedness between the PIR proteins in PvP01 versus Salvador-I (a), and the stability of the major clusters in several other P. vivax assemblies (b).
Panel a) presents a network illustrating the relatedness between the 1063 PIR proteins of PvP01 and 341 PIRs of Salvador-I (Sal-I) with length greater than 150 amino acids. The PvP01 PIRs are illustrated by black dots (nodes). The Sal-I PIRs are illustrated by coloured dots with colour-coding according to the subfamily classification of Lopez et al 23 as follows; purple = A, pink = B, pale green = C, red = D, pale blue = E, orange = G, green = H, blue = I, white = J, yellow = K , and grey = unassigned genes. Two nodes (PIRs) are connected if they have a global similarity of at least 25%. With the exception of a few proteins, the majority of Sal-I PIRs demonstrate clustering consistent with the classification of Lopez et al. Five new, interconnected clusters comprising previously unassigned Sal-I PIRs are denoted with a white “X”. In Panel b, a heat map summarises the number of PIRs assigned to the 27 major clusters (minimum 15 PIRs in total) in five geographically divergent P. vivax strains; PvP01 (Papua Indonesia), PvT01 (Thailand), PvC01 (Central China), Sal-I (El Salvador) and Brazil-I (Brazil). With the exception of Sal-I, which displayed fewer genes than the other isolates in several of the major clusters, the isolates demonstrated similar numbers of genes in most clusters.