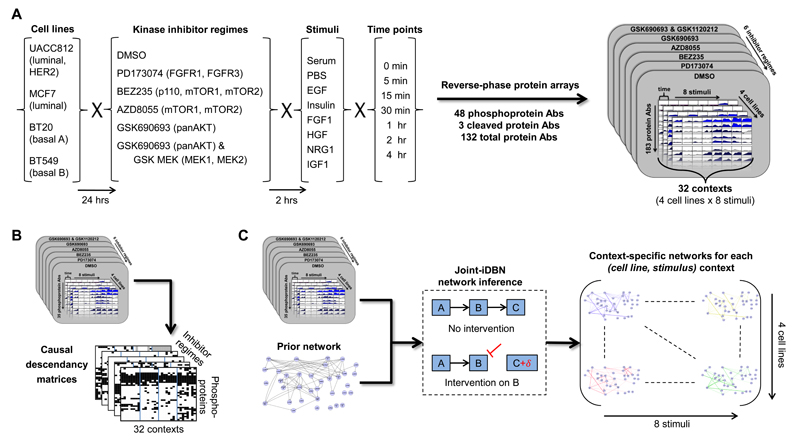
Figure 2. Data-Driven Reconstruction of Context-Specific Causal Signaling Networks.
(A) Overview of experimental approach. Reverse-phase protein arrays (RPPA) were used to investigate protein signaling in four human breast cancer cell lines under eight different stimuli. The combinations of cell line and stimulus defined 32 (cell line, stimulus) contexts. Prior to stimulus, cell lines were serum starved and treated with kinase inhibitors or DMSO control. RPPA assays were performed for each context at multiple time points post-stimulus, using more than 150 high-quality antibodies to target specific proteins, including approximately 40 phosphoproteins (the precise number of antibodies varies across cell lines; see STAR Methods and Table S1). (B) Causal descendancy matrices (CDMs). CDMs summarizing changes under intervention across all contexts were constructed for each intervention (see text for details). (C) Overview of causal network learning procedure. Interventional time-course data for each context were combined with existing biological knowledge in the form of a prior network to learn context-specific phosphoprotein signaling networks. Networks were learned using a variant of dynamic Bayesian networks designed for use with interventional data and that allowed joint learning over all 32 contexts at once (see STAR Methods).