Summary
Fungi, nematodes and oomycetes belong to the most prominent eukaryotic plant pathogenic organisms. Unicellular organisms from other eukaryotic lineages, commonly addressed as protists, also infect plants. This review provides an introduction to plant pathogenic protists, including algae infecting oomycetes, and their current state of research.
Keywords: algae, protist, plant pathogens, plasmodiophorids, stramenopiles, phytomonas, phytomyxae
Introduction
Molecular Plant Pathology has published a series of the Top 10 most important plant-pathogenic viruses (Scholthof et al., 2011), fungi (Dean et al., 2012), bacteria (Mansfield et al., 2012), nematodes (Jones et al., 2013) and oomycetes (Kamoun et al., 2015). The reviews of these major groups of plant pathogens do not cover a selection of protists that infect plants and algae leading to economically important diseases. These ‘non-standard’ plant pathogens are dispersed across the eukaryotic phylogenetic tree (Fig. 1), often in taxa unfamiliar to many plant pathologists as they are usually not associated with plant infections. In this review, we would like to introduce and raise awareness of such phylogenetically diverse eukaryotic plant pathogens.
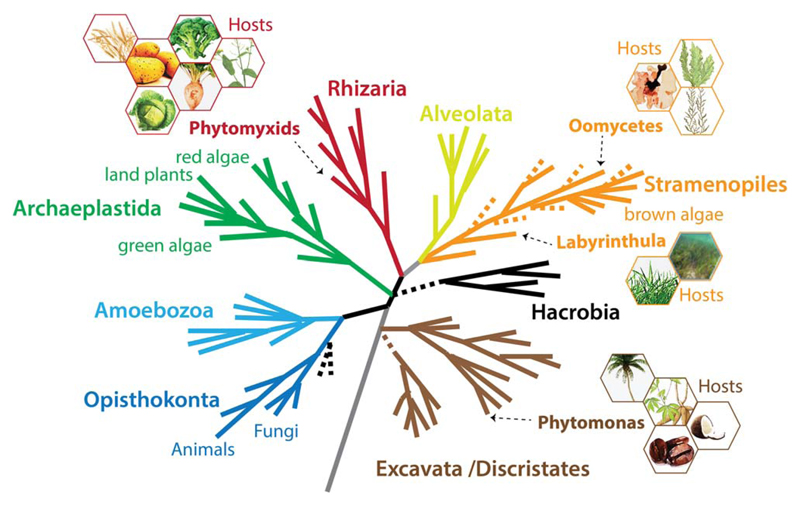
Fig. 1.
A schematic current eukaryotic tree of life indicating the phylogenetic positions of the eukaryotic plant pathogens outlined in this review. The hexagons show examples of the host species for each pathogen group. The phylogenetic tree was created by S. Baldauf (Uppsala University, Uppsala, Sweden) and reproduced with permission.
We describe diseases caused by these organisms, and the current state of research, especially with respect to their molecular biology and host interactions. We start with Phytomonas, plant pathogens in the trypanosomatids in the Excavata supergroup, a group better known as human and animal pathogens. They are followed by Phytomyxea, which are part of the Rhizaria supergroup and include agriculturally important plant pathogens, vectors of phytoviruses and species that infect marine plants and algae (Bulman and Braselton, 2014). Next, Labyrinthula are described, plant pathogens in the Stramenopiles, which are phylogenetic basal to oomycetes. Our review also includes marine oomycete parasites of red and brown algae, which impact on the fast growing aquaculture sector (Gachon et al., 2010). Advancing research in this field will benefit aquacultural sustainability and our understanding of higher oomycetes because of their basal phylogenetic position inside the oomycetes (Beakes et al., 2012).
Whole-genome or in-depth transcriptomic data for the species presented here are rare, with the exception of the Phytomyxea and Phytomonas. The organisms outlined reflect existing molecular knowledge; nevertheless, we emphasize that there are further important ‘unusual’ pathogens, especially on cultivated algae.
Excavata – Kinetoplastea Trypanosomatidae – Phytomonas
Trypanosomatids are a species-rich monophyletic group of obligate parasitic flagellates that are usually transmitted by insects. They are best known as agents of human and livestock diseases, such as sleeping sickness, Chagas disease and leishmaniosis, caused by Trypanosoma brucei, T. cruzi and Leishmania spp., respectively (Lukeš et al., 2014). Trypanosomatids also include the monophyletic genus Phytomonas (Fig. 2), which contains all known plant-dwelling trypanosomatids, some of which are pathogenic (Seward et al., 2016). The ancestral monoxenous lifestyle (development restricted to one host species) of trypanosomatids evolved at least three times independently into a dixenous strategy (Maslov et al., 2013) in Trypanosoma, Leishmania and Phytomonas (Lukeš et al., 2014). Phytomonas spp. are adapted to sap-sucking insects as primary hosts and plants as secondary hosts (Jaskowska et al., 2015). Phytomonas spp. were first described from the latex of Mediterranean spurge (Euphorbia pilulifera) (Lafont, 1909). Currently, the genus Phytomonas includes more than 200 species that colonize over 20 plant families (Camargo, 1999, Jaskowska et al., 2015).
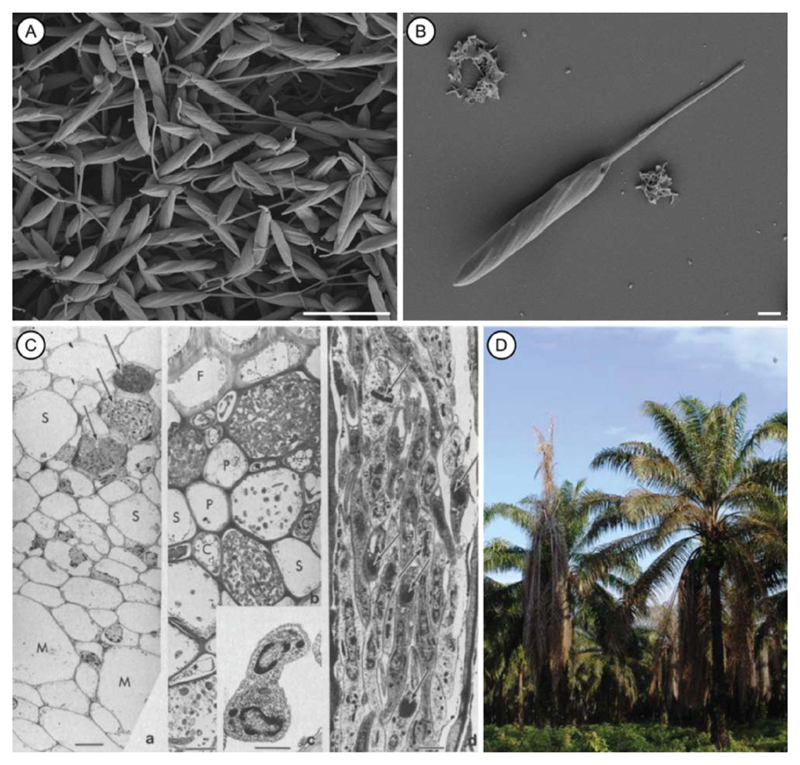
Fig. 2.
Phytomonas sp. and palm infections. (A, B) Scanning electron micrographs of Phytomonas serpens cells in culture (scale bars, 10 and 1 μm). (Courtesy of Martina Tesařová.) (C) Transmission electron micrographs of Phytomonas sp. flagellates in the phloem of coconut palms affected by hartrot. C, companion cell; F, fibre; M, immature metaxylem; P, phloem parenchyma cell; S, sieve elements free of flagellates. (a) Transverse section of a differentiating vascular bundle, showing recently matured sieve elements filled with flagellates (scale bar, 10 μm). (b) Transverse section of the phloem in palm with advanced symptoms (scale bar, 5 μm). (c) Transverse section of a dividing flagellate (scale bar, 0.5 μm). (d) Longitudinal section of a sieve element filled with flagellates. Arrows indicate the kinetoplast DNA (scale bar, 1 μm). (Reproduced from Parthasarathy et al., 1976.) (D) Coconut palms with symptoms of hartrot. (Photograph: Monica L. Elliott, Professor, Plant Pathology, University of Florida, Institute of Food and Agricultural Sciences (UF/IFAS), Gainesville, FL, USA.)
Phytomonas spp. can be separated into four ecological sub-groups based on whether they inhabit the latex ducts, fruits, phloem or flowers of their host plants (Camargo, 1999). Most commonly, Phytomonas spp. reside in latex ducts, yet the most pathogenic species are phloem dwelling, such as P. leptovasorum and P. staheli, which cause coffee phloem necrosis (CPN) and palm wilts, respectively (Jaskowska et al., 2015). Phytomonas leptovasorum infection triggers multiple divisions of the sieve tubes in coffee roots, leading to CPN. The disease is a potential threat to Brazil as the world’s largest coffee exporter, from which CPN has been reported, but never spread (Camargo, 1999). This disease occurs either acutely (plants wither and die within 2 months) or chronically (plants gradually die within a year) (Stahel, 1931).
Phytomonas staheli causes wilts of coconut (Cocos nucifera) and oil palms (Elaeis guineensis) (McGhee and McGhee, 1979). Both deadly wilts, ‘hartrot’ of coconut palms and ‘marchitez sorpresiva’ of oil palms, are characterized by progressive leaf browning, followed by rapid rotting of fruits, spears and roots (Kastelein, 1987; Lopez, 1975). Slow wilt of oil palms (‘marchitez lenta’) manifests as additional chlorosis (Di Lucca et al., 2013). Symptomless plants and wild hosts can harbour Phytomonas flagellates (Di Lucca et al., 2013). Potential disease outbreaks constantly threaten palm cultivation in South and Central America. In one Surinamese district, Phytomonas destroyed half of the coconut population (Kastelein, 1987). The latex-inhabiting P. françai is linked to empty roots disease (‘chochamento de raizes’) of the Unha cassava (Manihot esculenta) variety, although its pathogenicity remains unclear (Jaskowska et al., 2015; Kitajima et al., 1986).
The first Phytomonas draft genome came from the tomato fruit-inhabiting P. serpens (Kořený et al., 2012), which produces no significant systemic disease, but causes yellow spots on fruit (Camargo, 1999). The genomes of the pathogenic phloem-specific Phytomonas strain HART1 from Guyanan coconut and the non-symptomatic latex-specific strain EM1 from Euphorbia were generated shortly after (Porcel et al., 2014). Recently, the genome of the cassava latex-inhabiting P. françai has been announced (Butler et al., 2017), which will enable comparative genomics of Phytomonas spp. with different host and ecological lifestyles in the future.
The Phytomonas genomes are compact, consisting of single-copy genes, and are almost free of transposable elements and repeats. Therefore they are smaller (≅18 Mb) than most trypanosomatid genomes (26–33 Mb). Phytomonas spp. contain only about 6400 protein-coding genes versus approximately 10 400 found typically in trypanosomatids.
As in other biotrophs, Phytomonas metabolism is highly adapted to parasitic lifestyles. These plant pathogens contain fewer genes involved in amino acid synthesis and energy metabolism and fewer protein kinases than the related Leishmania and Trypanosoma spp. Fatty acids (FAs) are synthesized via elongases instead of de novo, as FA synthases are missing (Porcel et al., 2014). Phytomonas spp. have the unique capacity amongst trypanosomatids to live in the total absence of haem, although they might be able to scavenge it (Kořený et al., 2012). In addition, they have lost several cytochrome subunits of respiratory complexes. For energy production, Phytomonas may depend solely on glycolysis, whereas other trypanosomatids (at least in part of their life cycle) rely on mitochondrial amino acid metabolism as their main energy source (Jaskowska et al., 2015; Porcel et al., 2014). As their insect vector(s) feed on carbohydrate-rich plant juices, Phytomonas might not require a switch from carbohydrate to amino acid metabolism. Phytomonas spp. contain complete sets of glycolytic enzymes and large numbers of glycosomes, into which glycolysis is compartmentalized (Hannaert et al., 2003; Porcel et al., 2014). Also unique amongst trypanosomatids, Phytomonas spp. possess the capacity to feed on plant polysaccharides using glucoamylase and α-glucosidase enzymes. In addition, an α,α-trehalose phosphorylase, acquired by horizontal gene transfer, enables feeding on trehalose, a common sugar in the plant and insect hosts of Phytomonas (Porcel et al., 2014).
The Phytomonas HART1 and EM1 isolates share a majority of genes. However, only the phloem-restricted pathogenic HART1 encodes invertase genes for the degradation of sucrose (Porcel et al., 2014), probably as an adaptation to the abundance of sucrose in the phloem. For both the HART1 and EM1 isolates, 282 secreted proteins were predicted. Their secretomes contain no plant cell wall-degrading enzymes, which reflects the feeding of the pathogens on extracellular plant fluids. It is unknown whether Phytomonas spp. secrete protein effectors, which modulate host plant immune responses. However, several aspartyl proteases that are absent from the genomes of Leishmania and Trypanosoma are secreted in both Phytomonas strains (Porcel et al., 2014). These proteases may be involved in Phytomonas–host interactions, as seen for oomycete and fungal plant pathogens (Jashni et al., 2015). The pathogenic HART1 strain carries five copies of a cathepsin-like aspartyl protease, derived from duplication events, whereas EM1 has only a single copy. This implies that these enzymes are potential virulence factors (Porcel et al., 2014). The gene family of major surface proteases, which are involved in the pathogenicity of Leishmania, underwent an expansion in the genus Phytomonas (Jackson, 2015). The surface glycoprotein 63 subfamily is present in 20 copies in HART1 and only twice in EM1, a putative adaptation of HART1 to the phloem environment (Jaskowska et al., 2015; Porcel et al., 2014).
Although the procyclic stage of Phytomonas spp. can be easily cultivated, an experimental system including their plant host is not available. Hence, our understanding of how these plant-dwelling or plant-parasitizing flagellates interact with their plant hosts is only at an early stage.
Currently, there is no treatment or prevention of the diseases caused by Phytomonas, except for the simple extermination of infected plants (Jaskowska et al., 2015). However, it has been observed that the tomato (Solanum lycopersicum) is relatively resistant to P. serpens, as the parasite only causes yellow spots on its fruits, resulting in their lower commercial value. Interestingly, the tomato defensive alkaloids tomatine and tomatidine, surface-active saponin-like compounds, induce permeabilization and vacuolization of the parasite (Medina et al., 2015). Both alkaloids inhibit the growth of P. serpens and therefore represent potential therapeutic agents against these phytopathogens (Medina et al., 2015).
Rhizaria
Phytomyxea – plasmodiophorids
The obligate biotrophic Plasmodiophorida (plasmodiophorids) belong to the Phytomyxea (phytomyxids) in the eukaryotic supergroup Rhizaria (Fig. 1) (Adl et al., 2012; Burki and Keeling, 2014; Burki et al., 2010). These organisms infect a wide variety of hosts, including oomycetes and brown algae (Neuhauser et al., 2014). Plasmodiophorids cause substantial damage to crops, including brassicas (Plasmodiophora brassicae), potatoes (Spongospora subterranea) and as vectors of viruses to beets, peanut and monocots (e.g. maize, rice, sugarcane, wheat, sorghum) (Polymyxa spp.) and potatoes (S. subterranea).
The plasmodiophorid life cycle consists of two phases: a sporangial stage leading to short-lived zoospores, and a sporogenic stage leading to the formation of persistent resting spores (Figs 3–5). Resting spores give rise to biflagellate primary zoospores which inject their cellular contents into host cells via a ‘Rohr und Stachel’ (Aist and Williams, 1971) (Fig. 3), initiating the sporangial life cycle stage. Multinucleate plasmodia develop and produce (mitotic) secondary zoospores, which can infect host cells and develop sporogenic multinucleate plasmodia that mature into resting spores. In the sporogenic stage, gall-causing plasmodiophorids induce division and massive enlargement of host cells (for greater detail, see Bulman and Braselton, 2014).
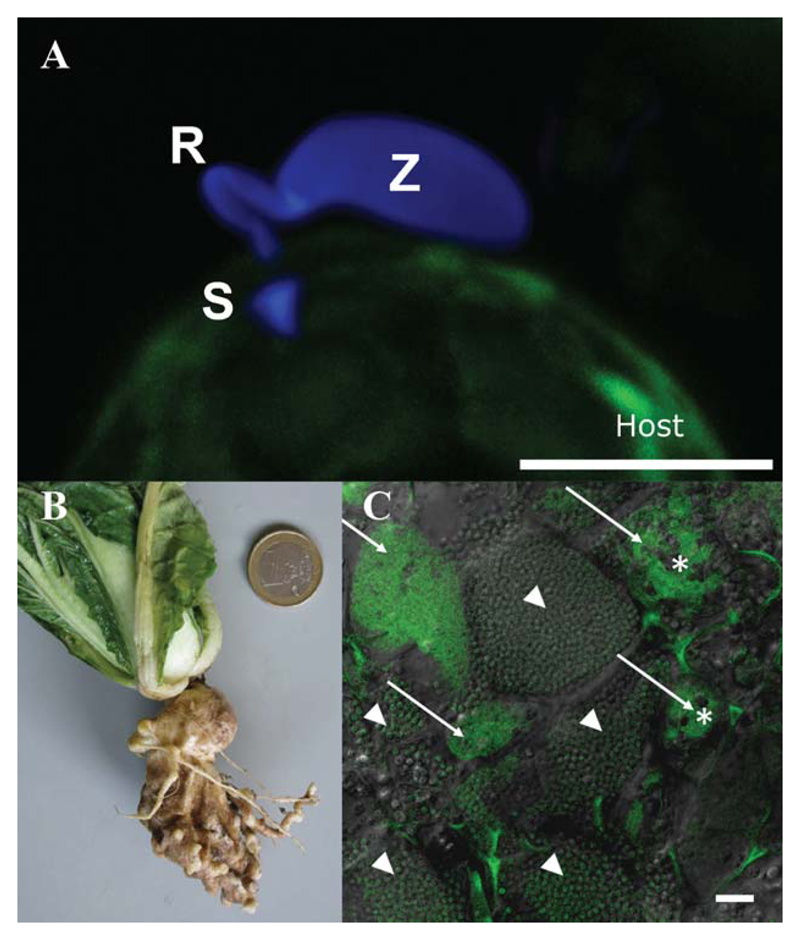
Fig. 3.
Phytomyxid infection and clubroot. (A) Phytomyxean parasites infect their host via a specialized extrusosome, called a ‘Rohr (R) and Stachel (S)’. The image shows a zoospore (Z) of the phagomyxid Maullinia ectocarpii infecting a female gametophyte of Macrocystis pyrifera (host). The M. ectocarpii spore was stained with calcofluor white and the host is visible via autofluorescence. Bar, 5 μm. (B) Clubroot symptoms on Chinese cabbage. (C) Laser scanning micrograph of Plasmodiophora brassicae resting spores (arrowheads) and plasmodia (arrows) in clubroot tissue. Plasmodia of different ages can be distinguished by the presence of typical vacuoles (asterisks), which disappear when the plasmodia start to differentiate into resting spores. Overlay of a light microscopic image and the signal of a Plasmodiophora-specific fluorescence in situ hybridization (FISH) probe (green: excitation, 488 nm; emission, 510–550 nm). Bar, 20 μm.
The durability of resting spores and inconsistent chemical control make the management of plasmodiophorid diseases difficult, and biological control efforts are only beginning (Ludwig-Müller, 2016; O’Brien and Milroy, 2017). Current management mostly relies on the use of resistant host varieties and crop rotation (Bittara et al., 2016; Ludwig-Müller, 2016). Pathogen detection and quantification in soil and in planta are important. Sequences of the ribosomal operon [i.e. 18S, 28S and internal transcribed spacer (ITS) ribosomal DNA (rDNA)] are widely used for these purposes (Bulman and Marshall, 1998; Faggian and Strelkov, 2009; van de Graaf et al., 2007; Vaianopoulos et al., 2007; Ward et al., 2004, 2005). Comparison of ITS and rDNA sequences has revealed various degrees of interspecific and intraspecific variation in plasmodiophorid species (Gau et al., 2013; van de Graaf et al., 2007; Qu and Christ, 2004; Schwelm et al., 2016).
Plasmodiophora brassicae
Plasmodiophora brassicae causes clubroot, a disease that leads to significant losses of Brassica oilseed and vegetable crop production worldwide (Dixon, 2009). Rapeseed cultivation for the production of biofuels, vegetable oils, industrial lubricants and rapeseed meal is of great economic importance, with a worldwide production of 27 million tonnes in 2012 (Carré and Pouzet, 2014). Clubroot has long been a major constraint for Brassica cultivation. A severe outbreak in 1872 in Russia led to the discovery of Pl. brassicae (Woronin, 1877). Clubroot causes crop losses of approximately 10% worldwide, but local losses are often greater (Dixon, 2009). Best practices for control are long crop rotation periods (although resting spores remain infective for decades), liming or cultivation of tolerant Brassica crops (Diederichsen et al., 2009; Ludwig-Müller, 2016). Clubroot resistance genes have been identified in Brassica genomes (Hatakeyama et al., 2013). However, resistance mechanisms are unclear and breakdown of ‘resistance’ has been repeatedly observed (Diederichsen et al., 2009; Strelkov et al., 2016; Zamani-Noor, 2017). Breeding for clubroot resistance is complicated as several pathotypes of Pl. brassicae exist. Genetic differences exist between Pl. brassicae strains, even within individual root galls, and chromosome polymorphism between strains has been suggested (Fähling et al., 2003; Graf et al., 2004; Klewer et al., 2001). However, molecular markers for Pl. brassicae pathotypes have yet to be established.
The genome of a European Pl. brassicae single-spore isolate has been generated recently (Schwelm et al., 2015), followed shortly after by genomic data for isolates from Canada and China (Bi et al., 2016; Rolfe et al., 2016). The Pl. brassicae genome is small (24.2–25.5 Mb), as a result of a high gene density and few repetitive elements (2%–5%) (Rolfe et al., 2016; Schwelm et al., 2015). The first single-nucleotide polymorphism (SNP) cluster analyses of the available Pl. brassicae genomes indicated relationships between SNPs, host ranges and regional origins (Rolfe et al., 2016). Additional genome sequencing of Pl. brassicae isolates should shed light on Pl. brassicae genomic diversity and pathotype-specific features.
The Pl. brassicae genomes show similar features to those of other biotrophic plant pathogens. Host dependence is evident, i.e. from a reduced number of biosynthesis genes for thiamine and certain amino acids (Rolfe et al., 2016; Schwelm et al., 2015). Transporter proteins may aid nutrient acquisition from the hosts (Rolfe et al., 2016). The Pl. brassicae genome encodes few carbohydrate-active enzymes (CAZymes). Genes encoding for plant cell wall-degrading enzymes are also rare, possibly a consequence of the mechanical penetration strategy via a ‘Rohr und Stachel’. However, chitin-related enzymes are enriched (Rolfe et al., 2016; Schwelm et al., 2015), which are probably involved in building the chitinous resting spore cell walls (Moxham and Buczacki, 1983).
In root galls, different life cycle stages of Pl. brassicae occur simultaneously (Fig. 3), making time course experiments difficult. The transcriptomics of isolated plasmodia show a highly active metabolism, i.e. the high expression of glyoxylate cycle-related genes suggests a high turnover from carbohydrates and lipids in the plasmodia (Schwelm et al. 2015). Lipids start to accumulate in the plasmodial stage and are stored in organelles in the plasmodia and resting spores (Bi et al., 2016; Moxham and Buczacki, 1983). The lipids are potential energy sources for resting spores and, as Pl. brassicae, like Phytomonas, does not contain an FA synthase (Schwelm et al., 2015), it might synthesize the lipids from host-derived precursors.
Depending on the strain sequenced, 553–590 secreted Pl. brassicae proteins were predicted. Effector candidates including the amino acid motif RxLR, known from Phytophthora effectors (Kamoun et al., 2015), are rare in Pl. brassicae (Rolfe et al., 2016; Schwelm et al., 2015). Crinkler (CRN)-related proteins were found in Pl. brassicae (Zhang et al., 2016a), but their functions are unknown. No effector candidates containing the chitin-binding LysM-motif, known to interfere with chitin detection in fungal-plant interactions (Kombrink and Thomma, 2013), were detected in Pl. brassicae.
Plasmodiophora brassicae infection results in a heavily altered host metabolism (Jubault et al., 2013): transcriptional and proteomic changes occur in pathways involved in lipid, flavonoid and plant hormone metabolism, defence responses, and carbohydrate and cell wall synthesis of the Brassica hosts (Agarwal et al., 2011; Chen et al., 2015, Ludwig-Müller et al., 2009; Päsold et al., 2010; Siemens et al., 2009; Zhang et al., 2016b). In Arabidopsis, gall formation results from increased host vascular cambium activity combined with significant reduction of xylem development (Malinowski et al., 2012). Conversely, higher activity of lignification-related genes occurs in less susceptible plants (Chen et al., 2015; Song et al., 2016).
On inoculation, amino acid transport and metabolism vary between tolerant and susceptible hosts, i.e. arginine and proline metabolism are less active in less susceptible B. rapa than in susceptible genotypes (Chen et al., 2015; Jubault et al., 2008; Song et al., 2016). Arginine and proline biosynthesis in Pl. brassicae also seems to be incomplete (Rolfe et al., 2016; Schwelm et al., 2015). Similar to other gall-forming plant diseases, galled roots also provoke hypoxic responses (Gravot et al., 2016). Infections by Pl. brassicae and morphogenic changes within roots leading to gall formation are accompanied by changes in phytohormone homeostasis of auxin, cytokinin and brassinosteroids (Agarwal et al., 2011; Ludwig-Müller et al., 2009; Schuller et al., 2014), but the exact mechanisms are not yet known. The contributions of plant hormones in clubroot have been addressed using Arabidopsis mutants altered in phytohormone biosynthesis, metabolism and signalling (Ludwig-Müller et al., 2017). In Arabidopsis, elevated cytokinins are associated with increased cell division early during infection. When galls are formed, the expression of host cytokinin biosynthetic genes is repressed, as is the expression of host cytokinin oxidases and dehydrogenases (Devos et al., 2006; Siemens et al., 2006). Plasmodiophora brassicae-produced cytokinins probably play a minor role in cytokinin homeostasis in infected tissues (Malinowski et al., 2016). Arabidopsis mutants of auxin conjugate synthesis, as well as auxin receptors, were more susceptible to the pathogen (Jahn et al., 2013), whereas nitrilase mutants were more tolerant (Grsic-Rausch et al., 2000). A Pl. brassicae protein can conjugate auxin and jasmonic acid to amino acids in vitro (Schwelm et al., 2015), but whether it manipulates host hormones in clubroots is unknown.
Effector-triggered immunity is likely to be important in host resistance to Pl. brassicae. During infection, resistance (R) genes and pathogen-related (PR) genes are expressed more strongly in tolerant than in susceptible plants, whereas the pathogen-associated molecular pattern (PAMP)-triggered immune response appears to be similar in both host types (Chen et al., 2015; Zhang et al., 2016b).
One Pl. brassicae effector candidate is a predicted secreted methyltransferase, PbBSMT. Biochemical expression assays have shown that this protein can mediate the methylation of salicylic acid (SA) (Ludwig-Müller et al., 2015). PbBSMT might interfere with SA signalling in infected root tissue. SA-mediated pathways are involved in resistance to Pl. brassicae (Agarwal et al., 2011; Lemarié et al., 2015; Lovelock et al., 2013). Accordingly, SA-responsive gene expression is increased in tolerant hosts (Chen et al., 2015; Song et al., 2016) and higher SA levels during early infection correlate with resistance (Chen et al., 2015; Zhang et al., 2016b).
Spongospora subterranea
Spongospora subterranea causes powdery scab of potato tubers (Solanum tuberosum) (Fig. 4A), an important blemish disease in most major potato-growing regions worldwide. This disease can result in the rejection of whole seed potato lots. The pathogen also causes root galling (Fig. 4B) and is the vector for the Potato mop top virus (PMTV, Pomovirus, Virgaviridae) (Merz and Falloon, 2009; Tamada and Kondo, 2013). Root membrane dysfunction, which reduces water uptake and plant growth, has also been attributed to S. subterranea (Falloon et al., 2016). All of these diseases devalue potato crops, causing potato tuber yield losses of >20% in severely diseased crops (Johnson and Cummings, 2015; Merz and Falloon, 2009; Shah et al., 2012). Mature tuber lesions and root galls are filled with clusters of resting spores (sporosori; Fig. 4D), each containing a primary zoospore. Root infection results in the development of zoosporangia (Fig. 4C) producing secondary zoospores. Both types of zoospore infect the host tuber, root epidermis cells and root hairs, and can transmit PMTV.
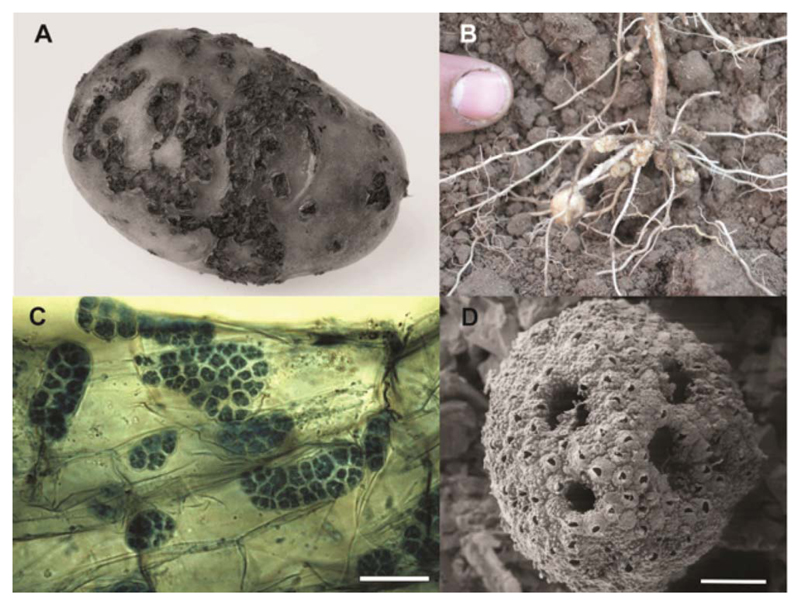
Fig. 4.
Potato infection by Spongospora subterranea. The potato pathogen Spongospora subterranea infects host tubers, roots and stolons, resulting in the development of powdery scab lesions (A) and galls (B). These usually appear in potato crops 2–3 months after planting, and mature to release sporosori (conglomerations of resting spores). A sporosorus contains 500–1000 resting spores, each containing a primary zoospore (D; bar, 10 μm). Secondary zoospores formed in zoosporangia (C; bar, 20 μm) emerge through root cell walls, disrupting host nutrient and water uptake.
Disease management is mainly preventative through the use of disease-free seed tubers and non-contaminated fields. Powdery scab and root galling susceptibility differ across potato cultivars (Bittara et al., 2016; Falloon et al., 2003), but no genetic basis of resistance has yet been identified. Metabolites of potato root exudates induce S. subterranea resting spore germination, but as l-glutamine and tyramine have the strongest effects, this might not be host specific (Balendres et al., 2016). This may explain reports of primary infection by S. subterranea in a range of non-solanaceous host plants (Merz and Falloon, 2009).
Spongospora subterranea ITS rDNA and microsatellite analyses indicate much greater genetic diversity in South American strains (the presumed origin of this pathogen) than elsewhere (Bulman and Marshall, 1998; Gau et al., 2013). After the initial dispersal from South America, Europe was probably the main source of spread of S. subterranea (Gau et al., 2013). Molecular data suggest possible substructures between root gall and tuber scab causing S. subterranea lineages from South America (Gau et al., 2013). Evidence for recombination in S. subterranea is limited, and there is little understanding of sexual recombination in phytomyxids (Bulman and Braselton, 2014).
Limited genomic sequences, including an assembled mitochondrial genome, are available from S. subterranea (Bulman et al., 2011; Gutiérrez et al., 2014, 2016). By comparison, relatively comprehensive S. subterranea transcriptomic datasets are available from root galls (Burki et al., 2010; Schwelm et al., 2015). As for Pl. brassicae, the current data suggest intron-rich genes, a paucity of CAZymes, but an enrichment of chitin-related enzymes in S. subterranea. By contrast, transposable elements are likely to be more common and expressed in S. subterranea than in Pl. brassicae (Bulman et al., 2011; Gutiérrez et al., 2014; Schwelm et al., 2015). For S. subterranean, 613 secreted proteins were predicted – enriched in ankyrin and protein domains – typical of effectors from other plant pathogens. Few are shared with Pl. brassicae, but a putative PbBSMT homologue was detected.
Although no genome has been published, genome sequences from S. subterranea are being generated. These will identify S. subterranea-specific features and allow research of the transcriptional interaction with its hosts.gg
Polymyxa spp
The genus Polymyxa includes two morphologically indistinguishable agriculturally important species: Polymyxa graminis (Fig. 5) and Polymyxa betae. Both differ in their rDNA sequences and host ranges. The host range of Px. betae is restricted to Chenopodiaceae and related plants, whereas Px. graminis infects mainly graminaceous plants (Legreve et al., 2000, 2002). Infection by these obligate root endoparasites is asymptomatic (Desoignies, 2012). Unlike Pl. brassicae and S. subterranea, Polymyxa spp. do not cause root galls on infected hosts, but indirectly cause damage as vectors of plant viruses. Polymyxa graminis transmits viruses belonging to Benyvirus, Bymovirus, Furovirus and Pecluvirus. They include economically important viruses of different grain crops, such as Barley yellow mosaic virus (BaYMV) and Soil-borne wheat mosaic virus (SBWMV), and also cause virus diseases on other cereals, sugar cane and peanuts [Peanut clump virus (PCV)] (Dieryck et al., 2011; Tamada and Kondo, 2013). Polymyxa betae transmits Beet necrotic yellow vein virus (BNYVV), causing ‘rhizomania’ in sugar beet (McGrann et al., 2009).
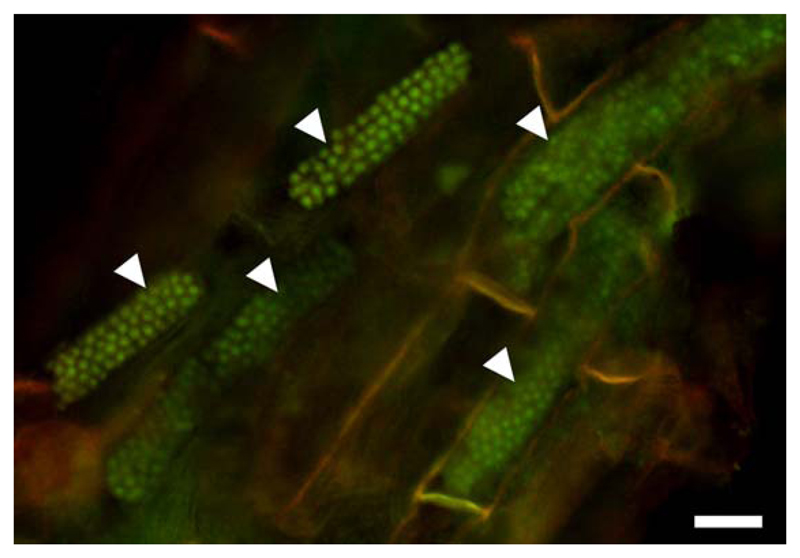
Fig. 5.
Resting spores of Polymyxa graminis in Poa sp. Resting spores are arranged in typical, long and cylindrical cytosori (arrowheads). The sample was stained with acridine orange, showing the nuclei of the fully developed resting spores. Epifluorescence micrograph obtained using blue excitation with long-pass emission (Nikon B-2A filter) allowing for the detection of DNA. Bar, 20 μm.
Polymyxa betae is a well-defined species, whereas, in Px. graminis, five formae speciales or six ribotypes exist, with subtype classifications based on ecological, molecular and biological characteristics, including specificity in virus transmission (Cox et al., 2014; Dieryck et al., 2011; Kanyuka et al., 2003; Legreve et al., 2002; Smith et al., 2013; Vaianopoulos et al., 2007; Ward et al., 2005; Ziegler et al., 2016).
Obtaining genomic data from Polymyxa spp. is more difficult than for the gall-forming plasmodiophorids as high-density infections with substantial amounts of parasite DNA cannot be identified. Polymyxa betae cultures on sugar beet hairy roots (Desoignies and Legreve, 2011) and in its non-natural host A. thaliana (Desoignies and Legreve, 2011; Smith et al., 2011) were tested, but were difficult to maintain. A suppression subtractive hybridization experiment identified most currently known Polymyxa gene models (Desoignies et al., 2014), including 76 Px. betae and 120 sugar beet expressed sequence tags (ESTs) putatively involved in the early stages of the host–pathogen interaction. The Px. betae ESTs included chitin synthase, polysaccharide deacetylases, ankyrins and galactose lectin domain-encoding transcripts, proteins which are also enriched in Pl. brassicae and S. subterranea (Bulman et al., 2011; Desoignies et al., 2014; Schwelm et al., 2015). Genes encoding for profilin and a von Willebrand factor domain-containing protein were also highly expressed. The sugar beet response to Px. betae infection, especially during the plasmodial stage, includes the over-expression of some defence genes, including those that encode PR proteins or lectins (Desoignies et al., 2014).
Other Phytomyxea
Other phytomyxids infect freshwater and marine organisms (Neuhauser et al., 2011). Maullinia ectocarpii (Fig. 3) and M. brasseltonii are plasmodiophorids infecting brown algae. Plasmodiophora diplantherea, Pl. bicaudata, Pl. halophile and Tetramyxa parasitica cause galls on seagrasses, and, in the case of T. parasitica, also other estuarine plants (Bulman and Braselton, 2014; Neuhauser et al., 2010). Spongospora nasturtii causes crook root on watercress and transmits the Watercress yellow spot virus (Walsh et al., 1989), impacting watercress cultivation.
Stramenopiles – Labyrinthula
Labyrinthula spp. are protists in the Labyrinthulida (Stramenopila), and are phylogenetically basal to oomycetes (Pan et al., 2017; Tsui et al., 2009). High-throughput environmental DNA sampling, ITS and ribosomal sequences suggest that Labyrinthula spp. are highly diverse, and globally distributed (Bockelmann et al., 2013; Collado-Mercado et al., 2010; Martin et al., 2016; Pan et al., 2017). These organisms are saline tolerant, and can be saprobes, coral inhabitants, endosymbionts of amoebae or endophytic facultative parasites of marine and terrestrial plants (Amon, 1978; Bigelow et al., 2005; Pan et al., 2017; Sullivan et al., 2013).
Marine Labyrinthula, such as L. zosterae, which causes seagrass wasting disease (SWD) (Sullivan et al., 2013), are usually associated with mangrove, macroalgal and seagrass ecosystems (Lindholm et al., 2016; Pan et al., 2017). Rapid blight disease (RBD) in turfgrasses is caused by the terrestrial species L. terrestris in high-salinity environments, such as salt lakes and golf course turf (Douhan et al., 2009; Kerrigan et al., 2012). This pathogen may have become important in specialized turfgrass because of increased salinity in irrigation or the use of reclaimed water, causing increased turf salinification (Olsen, 2007; Stowell et al., 2005). Both L. zosterae and L. terrestris vary greatly in virulence to their hosts (Chitrampalam et al., 2015; Douhan et al., 2009; Martin et al., 2016). Although the exact mechanism is uncertain, SWD and RBD manifest through the penetration of host leaf epidermis cells of individual Labyrinthula cells.
After infection, Labyrinthula spp. destroy the host chloroplasts and advance to neighbouring cells. This creates lesions, sometimes killing entire leaves or plants through interruption of photosynthesis (Fig. 6). These pathogens are therefore found on the edges of progressing infections rather than within the host lesions (Muehlstein, 1992; Olsen, 2007; Sullivan et al., 2016). They can be isolated from infected leaf tissues as they emerge from tissues plated onto serum seawater agar solutions (Fig. 6). The individual spindle- to oval-shaped Labyrinthula cells move through colonies of self-generated ectoplasmodic networks or ‘slimeways’, which are thought to originate from specialized organelles called bothrosomes. In conjunction with pseudopodium extension, a net-like tube is created within which the cells move. The movement of cells occurs through the utilization of an actomyosin system (Preston and King, 2005). The slimeways are also thought to aid nutrient absorption (Vishniac, 1955). Labyrinthula cells contain two vacuoles, thought to serve as excretory organs in the cell and may also regulate osmotic pressure, as their presence depends on the environmental salinity (Young, 1943).
Fig. 6.
Labyrinthula and disease symptoms. (A) Single fusiform cells of the unicellular Labyrinthulomycota Labyrinthula protist. (B) Labyrinthula cells emerging from a seagrass leaf on serum seawater agar. Cells move through colonies of self-generated ectoplasmodic networks or ‘slimeways’, a net-like tube within which Labyrinthula are able to move. (C) Symptoms of the seagrass wasting disease 4 days following the artificial infection of seagrass blades.
The seagrass–Labyrinthula pathosystem is the best-studied relationship for this group. Quantitative polymerase chain reaction (PCR) has shown that Labyrinthula spp. occur in most marine eel-grass populations in Europe, but pathogenic species may only cause disease when infection is coupled with host stress (Bockelmann et al., 2013; Brakel et al., 2014). However, the potential impact of SWD was observed in the 1930s, when Labyrinthula killed up to 90% of Zostera marina, the most abundant Northern Hemisphere seagrass (reviewed in Muehlstein, 1989; Sullivan et al., 2013). Seagrass meadows are ecologically rich and productive marine ecosystems, and important carbon sinks (Christianen et al., 2013; Fourqurean et al., 2012). They support commercial fish nurseries (Jackson et al., 2001) and influence bacterial pathogen populations (Lamb et al., 2017). Despite the important ecological and economic roles of their hosts, and widespread evidence of their cause of severe disease, research in Labyrinthula pathology is still under development.
Labyrinthula spp. tolerate high temperatures up to 28 °C, but, in tropical and subtropical seagrasses, increased temperature results in reduced virulence (Olsen and Duarte, 2015). Low salinity also inhibits Labyrinthula growth (Muehlstein et al., 1988), and so seagrass meadows in high-salinity waters may have an advantage compared with those in truly marine locations (Vergeer et al., 1995). The transcriptomic host response to a Labyrinthula infection of seagrasses includes the down-regulation of genes related to reactive oxygen species (ROS) and chitinases, whereas a phenolic acid synthesis gene is highly expressed (Brakel et al., 2014). Phenolic metabolites may produce ‘synergistic’ host benefits. Resistance to Labyrinthula is density dependent, and diseased leaves have enhanced phenolic metabolite concentrations and these may reduce host susceptibility to Labyrinthula (Groner et al., 2016; McKone and Tanner, 2009; Trevathan-Tackett et al., 2015). The first seagrass genome (of Z. marina) has been published recently (Olsen et al., 2016). As a host for Labyrinthula, this expands the ability to investigate the genetic and molecular interactions between Labyrinthula and seagrass, and to improve our understanding of this potentially devastating pathogen.
Stramenopiles – oomycetes as algal parasites
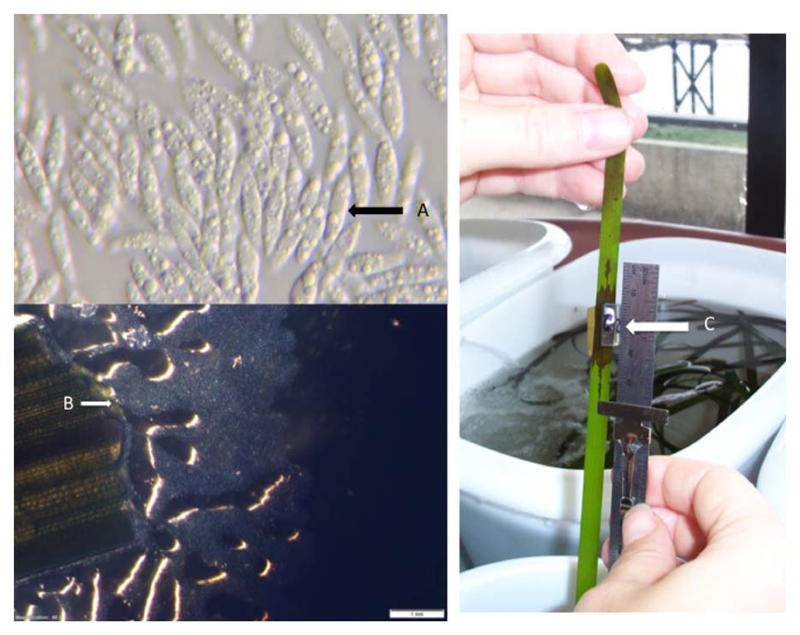
Oomycetes cause considerable damage in aquatic crops, including red (Rhodophyta) and brown (Phaeophyceae) algae. Worldwide algal industries have increased dramatically (Loureiro et al., 2015). In 2012, global macroalgal production was more than 23 million tonnes (dry weight), with a market value greater than six billion US$ (FAO, 2014). Most of this production (approximately 80%) is used for human consumption, and the remainder for fertilizers, animal feed additives and in medical and biotechnological applications, including biofuel production (Loureiro et al., 2015; Stengel and Connan, 2015). Seaweed farming is also often integrated into fish and shellfish aquaculture (Loureiro et al., 2015). The total market value for red seaweed reached 3.8 billion US$ (FAO, 2014). Best known in the form of Nori (sushi wrap), Pyropia (formerly Porphyra) spp. are the most common cultivated red algae. Brown algae are often the predominant primary producers in temperate and cold marine coastal ecosystems (Rodgers and Shears, 2016), and are phylogenetically distant from plants, green and red algae. They differ from red and green algae in cell wall composition (Michel et al., 2010), halogen metabolism (La Barre et al., 2010), oxylipin synthesis (Ritter et al., 2008) and life cycles (Coelho et al., 2011). Brown algae include edible seaweeds (e.g. kombu – Undaria pinnatifida, wakame – Saccharina japonica and sugar kelp – Saccharina latissima), and some species are commercially used to produce alginate. Collectively, red and brown algae are affected by many diseases (reviewed in Gachon et al., 2010). Because of the economic importance of Pyropia cultivation, and the growing economic burden of diseases for this crop (up to 50% of farm costs are spent on disease management: Kim et al., 2014), this review focuses on Pythium porphyrae and Olpidiopsis sp., the two main oomycetes that cause diseases on this crop.
Olpidiopsis diseases (previously ‘chytrid rot’) caused Korean Nori farms to lose nearly 25% of their resale value in 2012–2013 (Kim et al., 2014), but local losses can be greater (Klochkova et al., 2012; Loureiro et al., 2015). Environmental factors, such as temperature and seasonality, affect the severity of disease outbreaks.
Pythium porphyrae causes red rot disease, which is one of the most damaging diseases affecting Pyropia farming (Fig. 7) with production losses being greater than 20% (Kawamura et al., 2005). Distinct bleached patches on the algal blades characterize the initial infections. The diversity of Olpidiopsis is beginning to be described using molecular tools, with the recognition of new species, such as O. pyropiae from Korean farms (Klochkova et al., 2016; Sekimoto et al., 2008), in addition to the Japanese O. porphyrae.
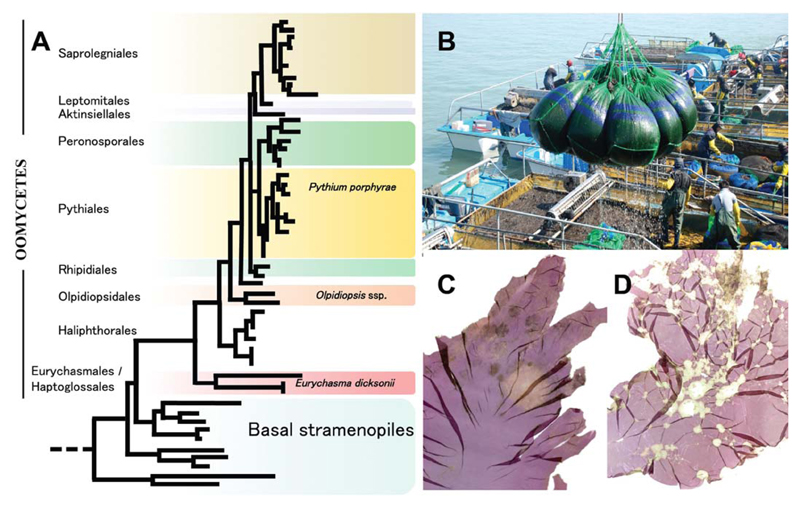
Fig. 7.
Oomycete phylogeny, Pyropia farming, Pythium porphyrae and Olpidiopsis symptoms. (A) Schematic phylogenetic tree of Oomycetes based on Beakes et al. (2012) indicating the positions of the discussed pathogens of marine algae. (B) Pyropia seaweed harvest on a commercial farm in South Korea (photograph: H. Kim). (C, D) Pyropia blade with lesions caused by Pythium porphyrae (C) and Olpidiopsis (D) infection. Photographs were originally published in Kim et al. (2014) which includes more detailed descriptions of Pyropia diseases.
Olpidiopsis spp
Olpidiopsis pathogens are obligate intracellular pathogens with biotrophic lifestyles. During the off-season of algal cultivation, Olpidiopsis may survive in alternative red algal hosts (e.g. Heterosiphonia sp.) or as dormant cysts (Klochkova et al., 2012, 2016). Germinating zoospores form germ tubes which penetrate algal cell walls. Within the cells, multinucleate walled thalli form, which quickly develop into sporangia, which release zoospores. With advancing infection, host cells break down and lesions in the blades become prominent.
The establishment of Olpidiopsis sp. and Pyropia pathosystems for research is difficult as the infected host disintegrates in a matter of days. However, with alternative hosts, such as Heterosiphonia japonica, stable dual cultures can be achieved (Klochkova et al., 2012). Olpidiopsis infection in this system is cell type specific, and occurs on the extended rhizoid-like apical cells. This specificity has been attributed to d(+)-mannose in host cell walls, indicating a specific lectin–carbohydrate interaction during host– parasite recognition, necessary for zoospore attachment to host cells (Klochkova et al., 2012). Until recently, the only available treatment for these diseases was to wash algal blades with acid, a practice now banned because of environmental concerns (Kim et al., 2014).
Pythium porphyrae
Red rot disease, caused by Py. porphyrae, was first described by Arasaki (1947). The disease spreads via zoospores and starts with distinct, small, red patches on the host blades in which the zoospores germinate. The pathogen develops extensive cell-to-cell spreading mycelium. Dead host cells change colour to violet–red and green before they degenerate, generating holes that finally destroy entire blades (Fig. 7). Red rot disease management is only effective during the early stages of infection, and PCR methods are important to detect the pathogen early during the algal cultivation period (Park et al., 2001, 2006). Disease control involves immersion of cultivation nets into organic acid, freezing of infected cultures and the application of fungicides (Amano et al., 1995; Hwang et al., 2009; Park et al., 2006). These treatments have significant costs and environmental impacts (Park and Hwang, 2015). Disease-resistant host cultivars are an alternative control strategy. Partially resistant Pyropia yezsoensis cultivars, generated from living cells in lesions of infected tissue, have altered cell wall polysaccharide contents (Park and Hwang, 2015). Sulfated galactans (e.g. porphyran) of the algal cell walls may be essential for cyst attachment and infection of Py. porphyrae, although the attraction and contact of zoospores are independent of host exudates (Uppalapati and Fujita, 2000). On resistant Pyropia sp., cysts with germ tubes frequently grow on the host thallus surfaces without penetration, and show no or delayed induction of appressoria (Uppalapati and Fujita, 2001). Although Py. porphyrae zoospores attach and encyst on a number of red algal species, red rot disease only develops on Pyropia yezoensis and Bangia atropurpurea (Uppalapati and Fujita, 2000).
Pythium porphyrae grows best in low-salinity water, possibly explaining why red rot occurs in farms near river banks (Klochkova et al., 2017). The pathogen can also infect and grow on land plants, including Chinese cabbage and rice. Pythium porphyrae carried from the land into coastal waters may increase damage in seaweed farms close to river inlets (Klochkova et al., 2017). This could enable molecular research on Py. porphyrae using the model hosts rice and A. thaliana. Genomic data are already available for Pyropia hosts (http://dbdata.rutgers.edu/nori/index.php) (Nakamura et al., 2013; Wu et al., 2014) and are currently being generated for Py. porphyrae and Olpidiopsis sp.
Eurychasma dicksonii
The most frequently recorded eukaryotic pathogen of brown algae is the biotrophic oomycete Eurychasma dicksonii. This phylogenetically basal oomycete (Beakes et al., 2012) is geographically widespread, tolerates a broad temperature range (4–23 8 °C) and infects at least 45 different species of brown algae in laboratory cultures (Müller et al., 1999). Similar to Olpidiopsis spp., Eu. dicksonii is a holocarpic endoparasite (Sekimoto et al. 2008). Zoospores attach, encyst and build adhesorium-like structures at the host surfaces. The parasite cytoplasm is transferred into the host via a needle-like structure which is associated with the formation of the adhesorium chamber at the host–spore contact point (Tsirigoti et al., 2015), similar to the plasmodiophorid ‘Rohr und Stachel’. After penetration, multinucleate non-walled immature thalli, with double membrane envelopes of host and parasite (Sekimoto et al., 2008), develop and expand in the infected host cells, until each cell is almost filled. The plasmodial thallus develops into a sporangium with peripheral primary cysts (Fig. 8), which release biflagellate zoospores through apical exit tubes. The empty cyst walls form a net-like sporangium, which is a distinctive morphological feature of this pathogen (Petersen, 1905).
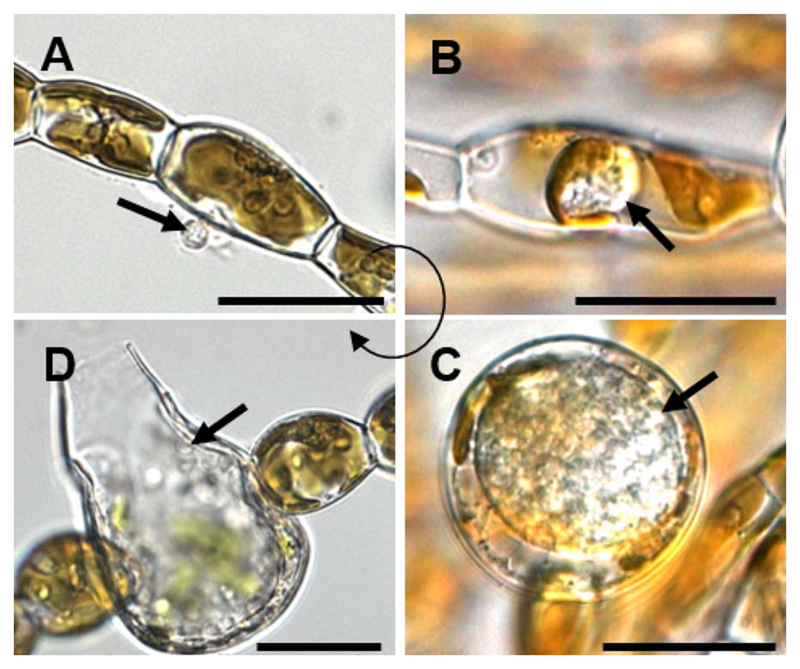
Fig. 8.
Life cycle of Eurychasma dicksonii in its brown algal host Ectocarpus siliculosus. (A) A spore (arrow) attaches to the algal surface and injects its content into the host. (B) Within the algal cytoplasm, the Eu. dicksonii thallus (arrow) develops which, at the early stage of infection, is unwalled. (C) Later, the pathogen thallus (arrow) has a cell wall and causes hypertrophic expansion of the algal host cell. (D) At the final stage, the complete thallus differentiates into a sporangium from which motile zoospores (arrow) are produced, completing the life cycle of the pathogen. Scale bars equal to 25 μm. (Figure reproduced from Strittmatter et al. 2016.)
Eurychasma dicksonii can be cultured in Ectocarpus siliculosus, the first brown alga to be genomically sequenced (Cock et al., 2010), explaining why the Eurychasma–Ectocarpus pathosystem is the most thoroughly investigated parasitic interaction in brown algae. A cDNA analysis of Ec. siliculosus infected with Eu. dicksonii identified 3086 unigenes of oomycete origin. The dataset of Eu. dicksonii included 351 proteins predicted to be secreted, but contained no CRN or RxLR effector candidates (Grenville-Briggs et al., 2011). The Eu. dicksonii genes included glucanases and a potential alginate lyase, for which no homologues in land plant-infecting oomycetes have been identified. Alginates and glucans are key components of brown algal cell walls. Similar to higher oomycetes, which secrete cell wall-degrading enzymes involved in host penetration, this lyase is probably an adaptation to the marine host. In brown algae, β-1,3-glucans are usually not part of the cell walls, but are storage polysaccharides. Cell wall modification is a putative host defence mechanism against Eu. dicksonii. On infection, cell wall thickening and increased amounts of β-1,3-glucans at the penetration site may build physical barriers to pathogen invasion. Large amounts of β-1,3-glucan occur at cell surfaces of partially resistant Ectocarpus strains (Tsirigoti et al., 2015).
Although the infection mechanisms remain largely unexplored, molecular data exist on the host response to infection by Eu. dicksonii. Host genes differentially expressed during infection include those encoding for proteins involved in the detoxification of ROS and halogen metabolism (Strittmatter et al., 2016). The host genome includes candidate immune receptors of the leucinerich and tetratricopeptide repeat families, which quickly evolve via an original exon shuffling mechanism (Zambounis et al., 2012). Different hosts display different levels of susceptibility to Eurychasma (Gachon et al., 2009), and the resistance mechanisms are currently being investigated using cytological and molecular approaches. A targeted movement of host nuclei to pathogen penetration sites has been observed (Grenville-Briggs et al., 2011), and microtubule disorganization in the host occurs only when zoosporogenesis of the pathogen begins (Tsirigoti et al., 2015).
Outlook
Our understanding of eukaryotic plant pathogens is built on studies of fungi, animals (both opisthokonts) and oomycetes (stramenopiles). For the plant pathogens introduced here, the biochemical interactions with their plant hosts are just beginning to be unravelled through the introduction of study systems (e.g. the Eu. dicksonii–brown algae interaction) or the generation of reference genomes (Pl. brassicae, Phytomonas spp.). This will allow the presented pathogens to take a more prominent place in the molecular plant pathology field in the coming years, create deeper insights into how these pathogens interact with their hosts and how they have evolved. This should finally lead to new strategies for the control of these pathogens.
Acknowledgements
A.S. was funded by Formas, the Swedish Research Council. S.N., J.B. and M.E. were funded by the Austrian Science Fund (grant Y0810-B16). S.B. and R.E.F. were funded by the New Zealand Ministry for Business Innovation and Employment (Programme LINX0804). J.L. was supported by the Czech Grant Agency award 15-21974 and the ERC CZ LL1601. We would like to thank Sandra Baldauf, Gwang Hoon Kim and Monica L. Elliott for providing the photographs used in the figures.
Footnotes
Author Contributions
A.S. and S.N. initiated and organized the manuscript, and the other authors are listed alphabetically. Section contributions are as follows: Phytomonas (J.L., A.N., A.S.), plasmodiophorids (A.S., S.N., S.B., R.E.F., U.M., N.D., A.L.), Labyrinthula (S.N., B.K.S.), oomycetes (C.M.M.G., M.S., A.S., S.N., J.B., M.E.). All the authors read the manuscript and agreed to publication.
The authors have no conflicts of interest to declare.
References
- Adl SM, Simpson AGB, Lane CE, Lukes J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, Heiss A, et al. The revised classification of eukaryotes. J Eukaryot Microbiol. 2012;59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A, Kaul V, Faggian R, Rookes JE, Ludwig-Muller J, Cahill DM. Analysis of global host gene expression during the primary phase of the Arabidopsis thaliana–Plasmodiophora brassicae interaction. Funct Plant Biol. 2011;38:462–478. doi: 10.1071/FP11026. [DOI] [PubMed] [Google Scholar]
- Aist JR, Williams PH. The cytology and kinetics of cabbage root hair penetration by Plasmodiophora brassicae. Can J Bot. 1971;49:2023–2034. [Google Scholar]
- Amano H, Suginaga R, Arashima K, Noda H. Immunological detection of the fungal parasite, Pythium sp. – the causative organism of red rot disease in Porphyra-yezoensis. J Appl Phycol. 1995;7:53–58. [Google Scholar]
- Amon JP. Thraustochytrids and labyrinthulids of terrestrial, aquatic and hypersaline environments of the Great Salt Lake, USA. Mycologia. 1978;70:1299–1301. [Google Scholar]
- Arasaki S. Studies on the rot of Porphyra tenera by Pythium. Nippon Suisan Gakkaishi. 1947;13:74–90. [Google Scholar]
- Balendres MA, Nichols DS, Tegg RS, Wilson CR. Metabolomes of potato root exudates: compounds that stimulate resting spore germination of the soil-borne pathogen Spongospora subterranea. J Agric Food Chem. 2016;64:7466–7474. doi: 10.1021/acs.jafc.6b03904. [DOI] [PubMed] [Google Scholar]
- Beakes GW, Glockling SL, Sekimoto S. The evolutionary phylogeny of the oomycete “fungi”. Protoplasma. 2012;249:3–19. doi: 10.1007/s00709-011-0269-2. [DOI] [PubMed] [Google Scholar]
- Bi K, He Z, Gao Z, Zhao Y, Fu Y, Cheng J, Xie J, Jiang D, Chen T. Integrated omics study of lipid droplets from Plasmodiophora brassicae. Sci Rep. 2016;6:36–965. doi: 10.1038/srep36965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigelow D, Olsen M, Gilbertson R. Labyrinthula terrestris sp. nov., a new pathogen of turf grass. Mycologia. 2005;97:185–190. doi: 10.3852/mycologia.97.1.185. [DOI] [PubMed] [Google Scholar]
- Bittara FG, Thompson AL, Gudmestad NC, Secor GA. Field evaluation of potato genotypes for resistance to powdery scab on tubers and root gall formation caused by Spongospora subterranea. Am J Potato Res. 2016;93:497–508. [Google Scholar]
- Bockelmann A-C, Tams V, Ploog J, Schubert PR, Reusch TB. Quantitative PCR reveals strong spatial and temporal variation of the wasting disease pathogen, Labyrinthula zosterae in northern European eelgrass (Zostera marina) beds. PLoS One. 2013;8:e62169. doi: 10.1371/journal.pone.0062169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakel J, Werner FJ, Tams V, Reusch TBH, Bockelmann AC. Current European Labyrinthula zosterae are not virulent and modulate seagrass (Zostera marina) defense gene expression. PLoS One. 2014;9:e92448. doi: 10.1371/journal.pone.0092448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulman S, Braselton JP. Rhizaria: Phytomyxea. In: McLaughlin DJ, Spatafora JW, editors. The Mycota VII, Part A, Systematics and Evolution. Springer; Berlin Heidelberg: 2014. pp. 99–112. [Google Scholar]
- Bulman S, Candy JM, Fiers M, Lister R, Conner AJ, Eady CC. Genomics of biotrophic, plant-infecting plasmodiophorids using in vitro dual cultures. Protist. 2011;162:449–461. doi: 10.1016/j.protis.2010.09.004. [DOI] [PubMed] [Google Scholar]
- Bulman SR, Marshall JW. Detection of Spongospora subterranea in potato tuber lesions using the polymerase chain reaction (PCR) Plant Pathol. 1998;47:759–766. [Google Scholar]
- Burki F, Keeling PJ. Rhizaria. Curr Biol. 2014;24:R103–R107. doi: 10.1016/j.cub.2013.12.025. [DOI] [PubMed] [Google Scholar]
- Burki F, Kudryavtsev A, Matz MV, Aglyamova GV, Bulman S, Fiers M, Keeling PJ, Pawlowski J. Evolution of Rhizaria: new insights from phylogenomic analysis of uncultivated protists. BMC Evol Biol. 2010;10:377. doi: 10.1186/1471-2148-10-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler CE, Jaskowska E, Kelly S. Genome sequence of Phytomonas françai a cassava (Manihot esculenta) latex parasite. Genome Announc. 2017;5:e01266–16. doi: 10.1128/genomeA.01266-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo EP. Phytomonas and other trypanosomatid parasites of plants and fruit. Adv Parasitol. 1999;42:29–112. doi: 10.1016/s0065-308x(08)60148-7. [DOI] [PubMed] [Google Scholar]
- Carré P, Pouzet A. Rapeseed market, worldwide and in Europe. OCL. 2014;21:D102. [Google Scholar]
- Chen J, Pang W, Chen B, Zhang C, Piao Z. Transcriptome analysis of Brassica rapa near-isogenic lines carrying clubroot-resistant and -susceptible alleles in response to Plasmodiophora brassicae during early infection. Front Plant Sci. 2015;6:1183. doi: 10.3389/fpls.2015.01183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitrampalam P, Goldberg N, Olsen MW. Labyrinthula species associated with turfgrasses in Arizona and New Mexico. Eur J Plant Pathol. 2015;143:485–493. [Google Scholar]
- Christianen MJA, van Belzen J, Herman PMJ, van Katwijk MM, Lamers LPM, van Leent PJM, Bouma TJ. Low-canopy seagrass beds still provide important coastal protection services. PLoS One. 2013;8:e62413. doi: 10.1371/journal.pone.0062413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock JM, Sterck L, Rouze P, Scornet D, Allen AE, Amoutzias G, Anthouard V, Artiguenave F, Aury JM, Badger JH, Beszteri B, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- Coelho SM, Godfroy O, Arun A, Le Corguillé G, Peters AF, Cock JM. Genetic regulation of life cycle transitions in the brown alga Ectocarpus. Plant Signal Behav. 2011;6:1858–1860. doi: 10.4161/psb.6.11.17737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Mercado E, Radway JC, Collier JL. Novel uncultivated labyrinthulomycetes revealed by 18S rDNA sequences from seawater and sediment samples. Aquat Microb Ecol. 2010;58:215–228. [Google Scholar]
- Cox BA, Luo H, Jones R. Polymyxa graminis isolates from Australia: identification in wheat roots and soil, molecular characterization and wide genetic diversity. Phytopathology. 2014;98:1567–1575. doi: 10.1094/PDIS-02-14-0128-RE. [DOI] [PubMed] [Google Scholar]
- Dean R, Van Kan JAL, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD. The Top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desoignies N. Polymyxa betae - Beta vulgaris: understanding the molecular interactions through transcriptome and plant defense analysis. PhD thesis; Université catholique de Louvain, Belgium: 2012. [Google Scholar]
- Desoignies N, Legreve A. In vitro dual culture of Polymyxa betae in Agrobacterium rhizogenes transformed sugar beet hairy roots in liquid media. J Eukaryot Microbiol. 2011;58:424–425. doi: 10.1111/j.1550-7408.2011.00563.x. [DOI] [PubMed] [Google Scholar]
- Desoignies N, Carbonell J, Moreau JS, Conesa A, Dopazo J, Legreve A. Molecular interactions between sugar beet and Polymyxa betae during its life cycle. Ann Appl Biol. 2014;164:244–256. [Google Scholar]
- Devos S, Laukens K, Deckers P, Van der Straeten D, Beeckman T, Inze D, Van Onckelen H, Witters E, Prinsen E. A hormone and proteome approach to picturing the initial metabolic events during Plasmodiophora brassicae infection on Arabidopsis. Mol Plant-Microbe Interact. 2006;19:1431–1443. doi: 10.1094/MPMI-19-1431. [DOI] [PubMed] [Google Scholar]
- Di Lucca AGT, Chipana EFT, Albujar MJT, Peralta WD, Piedra YCM, Zelada JLA. Slow wilt: another form of Marchitez in oil palm associated with trypanosomatids in Peru. Trop Plant Pathol. 2013;38:522–533. [Google Scholar]
- Diederichsen E, Frauen M, Linders EGA, Hatakeyama K, Hirai M. Status and perspectives of clubroot resistance breeding in crucifer crops. J Plant Growth Regul. 2009;28:265–281. [Google Scholar]
- Dieryck B, Weyns J, Doucet D, Bragard C, Legreve A. Acquisition and transmission of peanut clump virus by Polymyxa graminis on cereal species. Phytopathology. 2011;101:1149–1158. doi: 10.1094/PHYTO-12-10-0335. [DOI] [PubMed] [Google Scholar]
- Dixon GR. The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J Plant Growth Regul. 2009;28:194–202. [Google Scholar]
- Douhan GW, Olsen MW, Herrell A, Winder C, Wong F, Entwistle K. Genetic diversity of Labyrinthula terrestris, a newly emergent plant pathogen, and the discovery of new Labyrinthulid organisms. Mycol Res. 2009;113:1192–1199. doi: 10.1016/j.mycres.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Faggian R, Strelkov SE. Detection and measurement of Plasmodiophora brassicae. J Plant Growth Regul. 2009;28:282–288. [Google Scholar]
- Fähling M, Graf H, Siemens J. Pathotype separation of Plasmodiophora brassicae by the host plant. J Phytopathol. 2003;151:425–430. [Google Scholar]
- Falloon RE, Genet RA, Wallace AR, Butler RC. Susceptibility of potato (Solanum tuberosum) cultivars to powdery scab (caused by Spongospora subterranea f. sp subterranea), and relationships between tuber and root infection. Australas Plant Pathol. 2003;32:377–385. [Google Scholar]
- Falloon RE, Merz U, Butler RC, Curtin D, Lister RA, Thomas SM. Root infection of potato by Spongospora subterranea: knowledge review and evidence for decreased plant productivity. Plant Pathol. 2016;65:422–434. [Google Scholar]
- FAO. Food and Agriculture Organization of the United Nations. Fisheries and Aquaculture Information and Statistics Services. [accessed on Jul 26, 2014];2014 URL http://www.fao.org/figis/.
- Fourqurean JW, Duarte CM, Kennedy H, Marba N, Holmer M, Mateo MA, Apostolaki ET, Kendrick GA, Krause-Jensen D, McGlathery KJ, Serrano O. Seagrass ecosystems as a globally significant carbon stock. Nat Geosci. 2012;5:505–509. [Google Scholar]
- Gachon CM, Strittmatter M, Muller DG, Kleinteich J, Kupper FC. Detection of differential host susceptibility to the marine oomycete pathogen Eurychasma dicksonii by real-time PCR: not all algae are equal. Appl Environ Microbiol. 2009;75:322–328. doi: 10.1128/AEM.01885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon CM, Sime-Ngando T, Strittmatter M, Chambouvet A, Kim GH. Algal diseases: spotlight on a black box. Trends Plant Sci. 2010;15:633–640. doi: 10.1016/j.tplants.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Gau RD, Merz U, Falloon RE, Brunner PC. Global genetics and invasion history of the potato powdery scab pathogen, Spongospora subterranea f.sp subterranea. PLoS One. 2013;8:e67944. doi: 10.1371/journal.pone.0067944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Graaf P, Wale SJ, Lees AK. Factors affecting the incidence and severity of Spongospora subterranea infection and galling in potato roots. Plant Pathol. 2007;56:1005–1013. [Google Scholar]
- Graf H, Fähling M, Siemens J. Chromosome polymorphism of the obligate biotrophic parasite Plasmodiophora brassicae. J Phytopathol. 2004;152:86–91. [Google Scholar]
- Gravot A, Richard G, Lime T, Lemarié S, Jubault M, Lariagon C, Lemoine J, Vicente J, Robert-Seilaniantz A, Holdsworth MJ, Manzanares-Dauleux MJ. Hypoxia response in Arabidopsis roots infected by Plasmodiophora brassicae supports the development of clubroot. BMC Plant Biol. 2016;16:251. doi: 10.1186/s12870-016-0941-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenville-Briggs L, Gachon CMM, Strittmatter M, Sterck L, Kupper FC, van West P. A molecular insight into algal–oomycete warfare: cDNA analysis of Ectocarpus siliculosus infected with the basal oomycete Eurychasma dicksonii. PLoS One. 2011;6:e24500. doi: 10.1371/journal.pone.0024500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner ML, Burge CA, Kim CJS, Rees E, Van Alstyne KL, Yang S, Wyllie-Echeverria S, Harvell CD. Plant characteristics associated with wide-spread variation in eelgrass wasting disease. Dis Aquat Organ. 2016;118:159–168. doi: 10.3354/dao02962. [DOI] [PubMed] [Google Scholar]
- Grsic-Rausch S, Kobelt P, Siemens JM, Bischoff M, Ludwig-Müller J. Expression and localization of nitrilase during symptom development of the clubroot disease in Arabidopsis. Plant Physiol. 2000;122:369–378. doi: 10.1104/pp.122.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez P, Bulman S, Alzate JF, Ortíz MC, Marín M. Mitochondrial genome sequence of the potato powdery scab pathogen Spongospora subterranea. Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27:58–59. doi: 10.3109/19401736.2013.873898. [DOI] [PubMed] [Google Scholar]
- Gutiérrez PA, Alzate JF, Montoya MM. Analysis of carbohydrate metabolism genes of Spongospora subterranea using 454 pyrosequencing. Rev Fac Nal Agr Medellin. 2014;67:7247–7260. [Google Scholar]
- Hannaert V, Saavedra E, Duffieux F, Szikora JP, Rigden DJ, Michels PAM, Opperdoes FR. Plant-like traits associated with metabolism of Trypanosoma parasites. Proc Natl Acad Sci USA. 2003;100:1067–1071. doi: 10.1073/pnas.0335769100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama K, Suwabe K, Tomita RN, Kato T, Nunome T, Fukuoka H, Matsumoto S. Identification and characterization of Crr1a, a gene for resistance to clubroot disease (Plasmodiophora brassicae Woronin) in Brassica rapa L. PLoS One. 2013;8:e54745. doi: 10.1371/journal.pone.0054745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang EK, Park CS, Kakinuma M. Physicochemical responses of Pythium porphyrae (Oomycota), the causative organism of red rot disease in Porphyra to acidification. Aquacult Res. 2009;40:1777–1784. [Google Scholar]
- Jackson AP. Genome evolution in trypanosomatid parasites. Parasitology. 2015;142:S40–S56. doi: 10.1017/S0031182014000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Rowden AA, Attrill MJ, Bossey SJ, Jones MB. The importance of seagrass beds as a habitat for fishery species. Oceanogr Mar Biol. 2001;39:269–303. [Google Scholar]
- Jahn L, Mucha S, Bergmann S, Horn C, Staswick P, Steffens B, Siemens J, Ludwig-Müller J. The clubroot pathogen (Plasmodiophora brassicae) influences auxin signaling to regulate auxin homeostasis in Arabidopsis. Plants. 2013;2:726–749. doi: 10.3390/plants2040726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jashni MK, Mehrabi R, Collemare J, Mesarich CH, de Wit PJ. The battle in the apoplast: further insights into the roles of proteases and their inhibitors in plant–pathogen interactions. Front Plant Sci. 2015;6:584. doi: 10.3389/fpls.2015.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskowska E, Butler C, Preston G, Kelly S. Phytomonas: trypanosomatids adapted to plant environments. PLoS Pathog. 2015;11:e1004484. doi: 10.1371/journal.ppat.1004927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DA, Cummings TF. Effect of powdery scab root galls on yield of potato. Plant Dis. 2015;99:1396–1403. doi: 10.1094/PDIS-11-14-1170-RE. [DOI] [PubMed] [Google Scholar]
- Jones JT, Haegeman A, Danchin EGJ, Gaur HS, Helder J, Jones MG, Kikuchi T, Manzanilla-López R, Palomares-Rius JE, Wesemael WM, Perry RN. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol Plant Pathol. 2013;14:946–961. doi: 10.1111/mpp.12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubault M, Hamon C, Gravot A, Lariagon C, Delourme R, Bouchereau A, Manzanares-Dauleux MJ. Differential regulation of root arginine catabolism and polyamine metabolism in clubroot-susceptible and partially resistant Arabidopsis genotypes. Plant Physiol. 2008;146:2008–2019. doi: 10.1104/pp.108.117432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubault M, Lariagon C, Taconnat L, Renou J-P, Gravot A, Delourme R, Manzanares-Dauleux MJ. Partial resistance to clubroot in Arabidopsis is based on changes in the host primary metabolism and targeted cell division and expansion capacity. Funct Integr Genomics. 2013;13:191–205. doi: 10.1007/s10142-013-0312-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S, Furzer O, Jones JDG, Judelson HS, Ali GS, Dalio RJ, Roy SG, Schena L, Zambounis A, Panabières F, Cahill D. The Top 10 oomycete pathogens in molecular plant pathology. Mol Plant Pathol. 2015;16:413–434. doi: 10.1111/mpp.12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanyuka K, Ward E, Adams MJ. Polymyxa graminis and the cereal viruses it transmits: a research challenge. Mol Plant Pathol. 2003;4:393–406. doi: 10.1046/j.1364-3703.2003.00177.x. [DOI] [PubMed] [Google Scholar]
- Kastelein P. Investigations on ‘Hartrot’ of coconut and oilpalms in Suriname. PhD dissertation, Rijksuniversiteit te Utrecht; Netherlands: 1987. [Google Scholar]
- Kawamura Y, Yokoo K, Tojo M, Hishiike M. Distribution of Pythium porphyrae, the causal agent of red rot disease of Porphyrae spp., in the Ariake Sea, Japan. Plant Dis. 2005;89:1041–1047. doi: 10.1094/PD-89-1041. [DOI] [PubMed] [Google Scholar]
- Kerrigan JL, Olsen MW, Martin SB. Rapid blight of turfgrass. [accessed on Aug 1, 2017];Plant Health Instructor. 2012 https://www.apsnet.org/edcenter/intropp/lessons/fungi/other/Pages/RapidBlight.aspx. [Google Scholar]
- Kim GH, Moon KH, Kim JY, Shim J, Klochkova TA. A revaluation of algal diseases in Korean Pyropia (Porphyra) sea farms and their economic impact. Algae. 2014;29:249–265. [Google Scholar]
- Kitajima EW, Vainstein MH, Silveira JSM. Flagellate protozoan associated with poor development of the root-system of cassava in the Espirito-Santo State, Brazil. Phytopathology. 1986;76:638–642. [Google Scholar]
- Klewer A, Luerben H, Graf H, Siemens J. Restriction fragment length polymorphism markers to characterize Plasmodiophora brassicae single-spore isolates with different virulence patterns. J Phytopathol. 2001;149:121–127. [Google Scholar]
- Klochkova TA, Shim JB, Hwang MS, Kim GH. Host–parasite interactions and host species susceptibility of the marine oomycete parasite, Olpidiopsis sp., from Korea that infects red algae. J Appl Phycol. 2012;24:135–144. [Google Scholar]
- Klochkova TA, Shin YJ, Moon KH, Motomura T, Kim GH. New species of unicellular obligate parasite, Olpidiopsis pyropiae sp nov., that plagues Pyropia sea farms in Korea. J Appl Phycol. 2016;28:73–83. [Google Scholar]
- Klochkova TA, Jung S, Kim GH. Host range and salinity tolerance of Pythium porphyrae may indicate its terrestrial origin. J Appl Phycol. 2017;29:371–379. [Google Scholar]
- Kombrink A, Thomma BPHJ. LysM effectors: secreted proteins supporting fungal life. PLoS Pathog. 2013;9:e1003769. doi: 10.1371/journal.ppat.1003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kořený L, Sobotka RJK, Gnipová A, Flegontov P, Horváth A, Oborník M, Ayala FJ, Lukeš J. Aerobic kinetoplastid flagellate Phytomonas does not require heme for viability. Proc Natl Acad Sci USA. 2012;109:3808–3813. doi: 10.1073/pnas.1201089109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Barre S, Potin P, Leblanc C, Delage L. The halogenated metabolism of brown algae (Phaeophyta), its biological importance and its environmental significance. Mar Drugs. 2010;8:988–1010. doi: 10.3390/md8040988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafont A. Sur la présence d’un Leptomonas, parasite de la classe des Flagelles dans le lates de l’Euphorbia pilulifera. Cr séances Soc biol ses fil. 1909;66:1011–1013. [Google Scholar]
- Lamb JB, van de Water JAJM, Bourne DG, Altier C, Hein MY, Fiorenza EA, Abu N, Jompa J, Harvell CD. Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science. 2017;355:731–733. doi: 10.1126/science.aal1956. [DOI] [PubMed] [Google Scholar]
- Legreve A, Vanpee B, Delfosse P, Maraite H. Host range of tropical and sub-tropical isolates of Polymyxa graminis. Eur J Plant Pathol. 2000;106:379–389. [Google Scholar]
- Legreve A, Delfosse P, Maraite H. Phylogenetic analysis of Polymyxa species based on nuclear 5.8S and internal transcribed spacers ribosomal DNA sequences. Mycol Res. 2002;106:138–147. [Google Scholar]
- Lemarié S, Robert-Seilaniantz A, Lariagon C, Lemoine J, Marnet N, Jubault M, Manzanares-Dauleux MJ, Gravot A. Both the jasmonic acid and the salicylic acid pathways contribute to resistance to the biotrophic clubroot agent Plasmodiophora brassicae in Arabidopsis. Plant Cell Physiol. 2015;56:2158–2168. doi: 10.1093/pcp/pcv127. [DOI] [PubMed] [Google Scholar]
- Lindholm T, Lindqvist C, Sjöqvist C. Occurrence and activity of slime nets, Labyrinthula sp. among aquatic plants in cold and oligohaline Baltic Sea waters. Ann Bot Fennici. 2016;53:139–143. [Google Scholar]
- Lopez G, Genty P, Ollagnier M. Control preventivo de la “Marchitez sorpresiva” del Elaeis guineensis en America Latina. Oleagineux. 1975;30:243–250. [Google Scholar]
- Loureiro R, Gachon CM, Rebours C. Seaweed cultivation: potential and challenges of crop domestication at an unprecedented pace. New Phytol. 2015;206:489–492. doi: 10.1111/nph.13278. [DOI] [PubMed] [Google Scholar]
- Lovelock DA, Donald CE, Conlan XA, Cahill DM. Salicylic acid suppression of clubroot in broccoli (Brassicae oleracea var. italica) caused by the obligate biotroph Plasmodiophora brassicae. Australas Plant Pathol. 2013;42:141–153. [Google Scholar]
- Ludwig-Müller J. Belowground defence strategies against clubroot (Plasmodiophora brassicae) In: Vos CMF, Kazan K, editors. Belowground Defence Strategies in Plants. Cham: Springer International Publishing; 2016. pp. 195–219. [Google Scholar]
- Ludwig-Müller J, Prinsen E, Rolfe SA, Scholes JD. Metabolism and plant hormone action during clubroot disease. J Plant Growth Regul. 2009;28:229–244. [Google Scholar]
- Ludwig-Müller J, Jülke S, Geiß K, Richter F, Mithöfer A, Šola I, Rusak G, Keenan S, Bulman S. A novel methyltransferase from the intracellular pathogen Plasmodiophora brassicae methylates salicylic acid. Mol Plant Pathol. 2015;16:349–364. doi: 10.1111/mpp.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig-Müller J, Auer S, Jülke S, Marschollek S. Manipulation of auxin and cytokinin balance during the Plasmodiophora brassicae–Arabidopsis thaliana interaction. In: Dandekar T, Naseem M, editors. Auxins and Cytokinins in Plant Biology: Methods and Protocols. New York, NY: Springer; 2017. pp. 41–60. [DOI] [PubMed] [Google Scholar]
- Lukeš J, Skalicky T, Tyc J, Votypka J, Yurchenko V. Evolution of parasitism in kinetoplastid flagellates. Mol Biochem Parasitol. 2014;195:115–122. doi: 10.1016/j.molbiopara.2014.05.007. [DOI] [PubMed] [Google Scholar]
- Malinowski R, Smith JA, Fleming AJ, Scholes JD, Rolfe SA. Gall formation in clubroot-infected Arabidopsis results from an increase in existing meristematic activities of the host but is not essential for the completion of the pathogen life cycle. Plant J. 2012;71:226–238. doi: 10.1111/j.1365-313X.2012.04983.x. [DOI] [PubMed] [Google Scholar]
- Malinowski R, Novák O, Borhan MH, Spíchal L, Strnad M, Rolfe SA. The role of cytokinins in clubroot disease. Eur J Plant Pathol. 2016;145:543–557. [Google Scholar]
- Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P, Dow MAX, Verdier V, Beer SV, Machado MA, Toth IAN. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 2012;13:614–629. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DL, Chiari Y, Boone E, Sherman TD, Ross C, Wyllie-Echeverria S, Gaydos JK, Boettcher AA. Functional, phylogenetic and host-geographic signatures of Labyrinthula spp. provide for putative species delimitation and a global-scale view of seagrass wasting disease. Estuar Coasts. 2016;39:1–19. [Google Scholar]
- Maslov DA, Votypka J, Yurchenko V, Lukes J. Diversity and phylogeny of insect trypanosomatids: all that is hidden shall be revealed. Trends Parasitol. 2013;29:43–52. doi: 10.1016/j.pt.2012.11.001. [DOI] [PubMed] [Google Scholar]
- McGhee RB, McGhee AH. Biology and structure of Phytomonas staheli sp.n. a trypanosomatid located in sieve tubes of coconut and oil palms. J Protozool. 1979;26:348–351. [Google Scholar]
- McGrann GRD, Grimmer MK, Mutasa-Goettgens ES, Stevens M. Progress towards the understanding and control of sugar beet rhizomania disease. Mol Plant Pathol. 2009;10:129–141. doi: 10.1111/j.1364-3703.2008.00514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKone KL, Tanner CE. Role of salinity in the susceptibility of eelgrass Zostera marina to the wasting disease pathogen Labyrinthula zosterae. Mar Ecol Prog Ser. 2009;377:123–130. [Google Scholar]
- Medina JM, Rodrigues JCF, Moreira OC, Atella G, de Souza W, Barrabin H. Mechanisms of growth inhibition of Phytomonas serpens by the alkaloids tomatine and tomatidine. Mem Inst Oswaldo Cruz. 2015;110:48–55. doi: 10.1590/0074-02760140097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz U, Falloon RE. Review: powdery scab of potato—increased knowledge of pathogen biology and disease epidemiology for effective disease management. Potato Res. 2009;52:17–37. [Google Scholar]
- Michel G, Tonon T, Scornet D, Cock JM, Kloareg B. The cell wall polysaccharide metabolism of the brown alga Ectocarpus siliculosus. Insights into the evolution of extracellular matrix polysaccharides in Eukaryotes. New Phytol. 2010;188:82–97. doi: 10.1111/j.1469-8137.2010.03374.x. [DOI] [PubMed] [Google Scholar]
- Moxham SE, Buczacki ST. Chemical-composition of the resting spore wall of Plasmodiophora-brassicae. Trans Br Mycol Soc. 1983;80:297–304. [Google Scholar]
- Muehlstein LK. Perspectives on the wasting disease of eelgrass Zostera marina. Dis Aquat Organ. 1989;7:211–221. [Google Scholar]
- Muehlstein LK. The host–pathogen interaction in the wasting disease of eelgrass, Zostera marina. Can J Bot. 1992;70:2081–2088. [Google Scholar]
- Muehlstein LK, Porter D, Short FT. Labyrinthula sp, a marine slime-mold producing the symptoms of wasting disease in eelgrass, Zostera marina. Mar Biol. 1988;99:465–472. [Google Scholar]
- Müller DG, Küpper FC, Küpper H. Infection experiments reveal broad host ranges of Eurychasma dicksonii (Oomycota) and Chytridium polysiphoniae (Chytridiomycota), two eukaryotic parasites in marine brown algae (Phaeophyceae) Phycol Res. 1999;47:217–223. [Google Scholar]
- Nakamura Y, Sasaki N, Kobayashi M, Ojima N, Yasuike M, Shigenobu Y, Satomi M, Fukuma Y, Shiwaku K, Tsujimoto A, Kobayashi T. The first symbiont-free genome sequence of marine red alga, Susabi-nori (Pyropia yezoensis) PLoS One. 2013;8:e57122. doi: 10.1371/journal.pone.0057122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauser S, Bulman S, Kirchmair M. Plasmodiophorids: The Challenge to Understand Soil-Borne, Obligate Biotrophs with a Multiphasic Life Cycle. In: Gherbawy Y, Voigt K, editors. Molecular Identification of Fungi. Berlin, Heidelberg: Springer Berlin Heidelberg; 2010. pp. 51–78. [Google Scholar]
- Neuhauser S, Kirchmair M, Gleason FH. The ecological potentials of Phytomyxea (“plasmodiophorids”) in aquatic food webs. Hydrobiologia. 2011;659:23–35. doi: 10.1007/s10750-010-0508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauser S, Kirchmair M, Bulman S, Bass D. Cross-kingdom host shifts of phytomyxid parasites. BMC Evol Biol. 2014;14:33. doi: 10.1186/1471-2148-14-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien PA, Milroy SP. Towards biological control of Spongospora subterranea f. sp subterranea, the causal agent of powdery scab in potato. Australas Plant Pathol. 2017;46:1–10. [Google Scholar]
- Olsen JL, Rouze P, Verhelst B, Lin YC, Bayer T, Collen J, Dattolo E, De Paoli E, Dittami S, Maumus F, Michel G. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature. 2016;530:331–335. doi: 10.1038/nature16548. [DOI] [PubMed] [Google Scholar]
- Olsen MW. Labyrinthula terrestris: a new pathogen of cool-season turfgrasses. Mol Plant Pathol. 2007;8:817–820. doi: 10.1111/j.1364-3703.2007.00425.x. [DOI] [PubMed] [Google Scholar]
- Olsen YS, Duarte CM. Combined effect of warming and infection by Labyrinthula sp. on the Mediterranean seagrass Cymodocea nodosa. Mar Ecol Prog Ser. 2015;532:101–109. [Google Scholar]
- Pan JW, del Campo J, Keeling PJ. Reference tree and environmental sequence diversity of Labyrinthulomycetes. J Eukaryot Microbiol. 2017;64:88–96. doi: 10.1111/jeu.12342. [DOI] [PubMed] [Google Scholar]
- Park CS, Hwang EK. Biochemical characterization of Pyropia yezoensis-AP1 strain accompanies the resistance reaction to the red rot disease pathogen, Pythium porphyrae. J Appl Phycol. 2015;27:2149–2156. [Google Scholar]
- Park CS, Kakinuma M, Amano H. Detection and quantitative analysis of zoospores of Pythium porphyrae, causative organism of red rot disease in Porphyra, by competitive PCR. J Appl Phycol. 2001;13:433–441. [Google Scholar]
- Park CS, Kakinuma M, Amano H. Forecasting infections of the red rot disease on Porphyra yezoensis Ueda (Rhodophyta) cultivation farms. J Appl Phycol. 2006;18:295–299. [Google Scholar]
- Parthasarathy MV, Van Slobbe WG, Soudant C. Trypanosomatid flagellate in the phloem of diseased coconut palms. Science. 1976;192:1346–1348. doi: 10.1126/science.192.4246.1346. [DOI] [PubMed] [Google Scholar]
- Päsold S, Siegel I, Seidel C, Ludwig-Müller J. Flavonoid accumulation in Arabidopsis thaliana root galls caused by the obligate biotrophic pathogen Plasmodiophora brassicae. Mol Plant Pathol. 2010;11:545–562. doi: 10.1111/j.1364-3703.2010.00628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen HE. Contributions a la connaissance des Phycomycetes marines. Overs K Danske Vidensk Selsk Forh. 1905;5:439–488. [Google Scholar]
- Porcel BM, Denoeud F, Opperdoes F, Noel B, Madoui MA, Hammarton TC, Field MC, Da Silva C, Couloux A, Poulain J, Katinka M. The streamlined genome of Phytomonas spp. relative to human pathogenic kinetoplastids reveals a parasite tailored for plants. PLoS Genet. 2014;10:e1004007. doi: 10.1371/journal.pgen.1004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston TM, King CA. Actin-based motility in the net slime mould Labyrinthula: evidence for the role of myosin in gliding movement. J Eukaryot Microbiol. 2005;52:461–475. doi: 10.1111/j.1550-7408.2005.00064.x. [DOI] [PubMed] [Google Scholar]
- Qu XS, Christ BJ. Genetic variation and phylogeny of Spongospora subterranea f.sp subterranea based on ribosomal DNA sequence analysis. Am J Potato Res. 2004;81:385–394. [Google Scholar]
- Ritter A, Goulitquer S, Salaun JP, Tonon T, Correa JA, Potin P. Copper stress induces biosynthesis of octadecanoid and eicosanoid oxygenated derivatives in the brown algal kelp Laminaria digitata. New Phytol. 2008;180:809–821. doi: 10.1111/j.1469-8137.2008.02626.x. [DOI] [PubMed] [Google Scholar]
- Rodgers KL, Shears NT. Modelling kelp forest primary production using in situ photosynthesis, biomass and light measurements. Mar Ecol Prog Ser. 2016;553:67–79. [Google Scholar]
- Rolfe SA, Strelkov SE, Links MG, Clarke WE, Robinson SJ, Djavaheri M, Malinowski R, Haddadi P, Kagale S, Parkin IA, Taheri A. The compact genome of the plant pathogen Plasmodiophora brassicae is adapted to intracellular interactions with host Brassica spp. BMC Genomics. 2016;17:1–15. doi: 10.1186/s12864-016-2597-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof KBG, Adkins S, Czosnek H, Palukaitis P, Jacquot E, Hohn T, Hohn B, Saunders K, Candresse T, Ahlquist P, Hemenway C. Top 10 plant viruses in molecular plant pathology. Mol Plant Pathol. 2011;12:938–954. doi: 10.1111/j.1364-3703.2011.00752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller A, Kehr J, Ludwig-Müller J. Laser microdissection coupled to transcriptional profiling of Arabidopsis roots inoculated by Plasmodiophora brassicae indicates a role for brassinosteroids in clubroot formation. Plant Cell Physiol. 2014;55:392–411. doi: 10.1093/pcp/pct174. [DOI] [PubMed] [Google Scholar]
- Schwelm A, Fogelqvist J, Knaust A, Jülke S, Lilja T, Bonilla-Rosso G, Karlsson M, Shevchenko A, Dhandapani V, Choi SR, Kim HG. The Plasmodiophora brassicae genome reveals insights in its life cycle and ancestry of chitin synthases. Sci Rep. 2015;5:11153. doi: 10.1038/srep11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwelm A, Berney C, Bass D, Dixelius C, Neuhauser S. The large subunit rDNA sequence of Plasmodiophora brassicae does not contain intraspecies polymorphism. Protist. 2016;167:544–554. doi: 10.1016/j.protis.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekimoto S, Beakes GW, Gachon CMM, Muller DG, Kupper FC, Honda D. The development, ultrastructural cytology, and molecular phylogeny of the basal oomycete Eurychasma dicksonii, infecting the filamentous phaeophyte algae Ectocarpus siliculosus and Pylaiella littoralis. Protist. 2008;159:299–318. doi: 10.1016/j.protis.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Seward EA, Votypka J, Kment P, Lukeš J, Kelly S. Description of Phytomonas oxycareni n. sp. from the salivary glands of Oxycarenus lavaterae. Protist. 2016;168:71–79. doi: 10.1016/j.protis.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Shah FA, Falloon RE, Butler RC, Lister RA. Low amounts of Spongospora subterranea sporosorus inoculum cause severe powdery scab, root galling and reduced water use in potato (Solanum tuberosum) Australas Plant Pathol. 2012;41:219–228. [Google Scholar]
- Siemens J, Keller I, Sarx J, Kunz S, Schuller A, Nagel W, Schmülling T, Parniske M, Ludwig-Müler J. Transcriptome analysis of Arabidopsis clubroots indicate a key role for cytokinins in disease development. Mol Plant–Microbe Interact. 2006;19:480–494. doi: 10.1094/MPMI-19-0480. [DOI] [PubMed] [Google Scholar]
- Siemens J, Bulman S, Rehn F, Sundelin T. Molecular biology of Plasmodiophora brassicae. J Plant Growth Regul. 2009;28:245–251. [Google Scholar]
- Smith MJ, Adams MJ, Ward E. Evidence that Polymyxa species may infect Arabidopsis thaliana. FEMS Microbiol Lett. 2011;318:35–40. doi: 10.1111/j.1574-6968.2011.02236.x. [DOI] [PubMed] [Google Scholar]
- Smith MJ, Adams MJ, Ward E. Ribosomal DNA analyses reveal greater sequence variation in Polymyxa species than previously thought and indicate the possibility of new ribotype-host–virus associations. Environ Microbiol Rep. 2013;5:143–150. doi: 10.1111/1758-2229.12026. [DOI] [PubMed] [Google Scholar]
- Song T, Chu M, Lahlali R, Yu F, Peng G. Shotgun label-free proteomic analysis of clubroot (Plasmodiophora brassicae) resistance conferred by the gene Rcr1 in Brassica rapa. Front Plant Sci. 2016;7:1013. doi: 10.3389/fpls.2016.01013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahel G. Zur Kenntnis der Siebröhrenkrankheit (Phloemnekrose) des Kaffee-baumes in Surinam. II. Phytopathol Z. 1931;4:539–548. [Google Scholar]
- Stengel DB, Connan S. Marine algae: a source of biomass for biotechnological applications. In: Stengel DB, Connan S, editors. Natural Products from Marine Algae: Methods and Protocols. New York, NY: Springer; 2015. pp. 1–37. [DOI] [PubMed] [Google Scholar]
- Stowell LJ, Martin SB, Olsen MW, Bigelow D, Kohout M, Peterson PD, Camberato J, Gelernter WD. Rapid blight: a new plant disease. [accessed on Aug 1, 2017];APSnet Features. 2005 Jul; Available at: http://www.apsnet.org/publications/apsnetfeatures/Pages/RapidBlight.aspx. [Google Scholar]
- Strelkov SE, Hwang SF, Manolii VP, Cao T, Feindel D. Emergence of new virulence phenotypes of Plasmodiophora brassicae on canola (Brassica napus) in Alberta, Canada. Eur J Plant Pathol. 2016;145:517–529. [Google Scholar]
- Strittmatter M, Grenville-Briggs LJ, Breithut L, van West P, Gachon CMM, Kupper FC. Infection of the brown alga Ectocarpus siliculosus by the oomycete Eurychasma dicksonii induces oxidative stress and halogen metabolism. Plant Cell Environ. 2016;39:259–271. doi: 10.1111/pce.12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan BK, Sherman TD, Damare VS, Lilje O, Gleason FH. Potential roles of Labyrinthula spp. in global seagrass population declines. Fungal Ecol. 2013;6:328–338. [Google Scholar]
- Sullivan BK, Robinson KL, Trevathan-Tackett SM, Lilje ES, Gleason FH, Lilje O. The first isolation and characterisation of the protist Labyrinthula sp. in Southeastern Australia. J Eukaryot Microbiol. 2016;64:504–513. doi: 10.1111/jeu.12387. [DOI] [PubMed] [Google Scholar]
- Tamada T, Kondo H. Biological and genetic diversity of plasmodiophorid-transmitted viruses and their vectors. J Gen Plant Pathol. 2013;79:307–320. [Google Scholar]
- Trevathan-Tackett SM, Lane AL, Bishop N, Ross C. Metabolites derived from the tropical seagrass Thalassia testudinum are bioactive against pathogenic Labyrinthula sp. Aquat Bot. 2015;122:1–8. [Google Scholar]
- Tsirigoti A, Beakes GW, Herve C, Gachon CM, Katsaros C. Attachment, penetration and early host defense mechanisms during the infection of filamentous brown algae by Eurychasma dicksonii. Protoplasma. 2015;252:845–856. doi: 10.1007/s00709-014-0721-1. [DOI] [PubMed] [Google Scholar]
- Tsui CK, Marshall W, Yokoyama R, Honda D, Lippmeier JC, Craven KD, Peterson PD, Berbee ML. Labyrinthulomycetes phylogeny and its implications for the evolutionary loss of chloroplasts and gain of ectoplasmic gliding. Mol Phylogenet Evol. 2009;50:129–140. doi: 10.1016/j.ympev.2008.09.027. [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Fujita Y. Carbohydrate regulation of attachment, encystment, and appressorium formation by Pythium porphyrae (Oomycota) zoospores on Porphyra yezoensis (Rhodophyta) J Phycol. 2000;36:359–366. [Google Scholar]
- Uppalapati SR, Fujita Y. The relative resistances of Porphyra species (Bangiales, Rhodophyta) to infection by Pythium porphyrae (Peronosporales, Oomycota) Bot Mar. 2001;44:1–7. [Google Scholar]
- Vaianopoulos C, Bragard C, Moreau V, Maraite H, Legreve A. Identification and quantification of Polymyxa graminis f. sp temperata and P. graminis f. sp tepida on barley and wheat. Plant Dis. 2007;91:857–864. doi: 10.1094/PDIS-91-7-0857. [DOI] [PubMed] [Google Scholar]
- Vergeer LHT, Aarts TL, Degroot JD. The wasting disease and the effect of abiotic factors (light-intensity, temperature, salinity) and infection with Labyrinthula zosterae on the phenolic content of Zostera marina shoots. Aquat Bot. 1995;52:35–44. [Google Scholar]
- Vishniac HS. The nutritional requirements of isolates of Labyrinthula spp. J Gen Microbiol. 1955;12:455–463. doi: 10.1099/00221287-12-3-455. [DOI] [PubMed] [Google Scholar]
- Walsh JA, Clay CM, Miller A. A new virus disease of watercress in England. EPPO Bull. 1989;19:463–470. [Google Scholar]
- Ward E, Kanyuka K, Motteram J, Kornyukhin D, Adams MJ. The use of conventional and quantitative real-time PCR assays for Polymyxa graminis to examine host plant resistance, inoculum levels and intraspecific variation. New Phytol. 2005;165:875–885. doi: 10.1111/j.1469-8137.2004.01291.x. [DOI] [PubMed] [Google Scholar]
- Ward LI, Fenn MGE, Henry CM. A rapid method for direct detection of Polymyxa DNA in soil. Plant Pathol. 2004;53:485–490. [Google Scholar]
- Woronin M. Plasmodiophora brassicae, der Organismus, der die unter dem Namen Hernie bekannte Krankheit der Kohlpflanzen verursacht. Arb St Petersburger naturf Ges. 1877;8:169–201. [Google Scholar]
- Wu S, Sun J, Chi S, Wang L, Wang X, Liu C, Li X, Yin J, Liu T, Yu J. Transcriptome sequencing of essential marine brown and red algal species in China and its significance in algal biology and phylogeny. Acta Oceanol Sin. 2014;33:1–12. [Google Scholar]
- Young EL. Studies on Labyrinthula. The etiologic agent of the wasting disease of eel-grass. Am J Bot. 1943;30:586–593. [Google Scholar]
- Zamani-Noor N. Variation in pathotypes and virulence of Plasmodiophora brassicae populations in Germany. Plant Pathol. 2017;66:316–324. [Google Scholar]
- Zambounis A, Elias M, Sterck L, Maumus F, Gachon CM. Highly dynamic exon shuffling in candidate pathogen receptors … what if brown algae were capable of adaptive immunity? Mol Biol Evol. 2012;29:1263–1276. doi: 10.1093/molbev/msr296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DP, Burroughs AM, Vidal ND, Iyer LM, Aravind L. Transposons to toxins: the provenance, architecture and diversification of a widespread class of eukaryotic effectors. Nucleic Acids Res. 2016a;44:3513–3533. doi: 10.1093/nar/gkw221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Liu Y, Fang Z, Li Z, Yang L, Zhuang M, Zhang Y, Lv H. Comparative transcriptome analysis between Broccoli (Brassica oleracea var. italica) and wild cabbage (Brassica macrocarpa Guss.) in response to Plasmodiophora brassicae during different infection stages. Front Plant Sci. 2016b;7:1929. doi: 10.3389/fpls.2016.01929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler A, Fomitcheva V, Zakri AM, Kastirr U. Occurrence of Polymyxa graminis ribotypes in Germany and their association with different host plants and viruses. Cereal Res Commun. 2016;44:251–262. [Google Scholar]