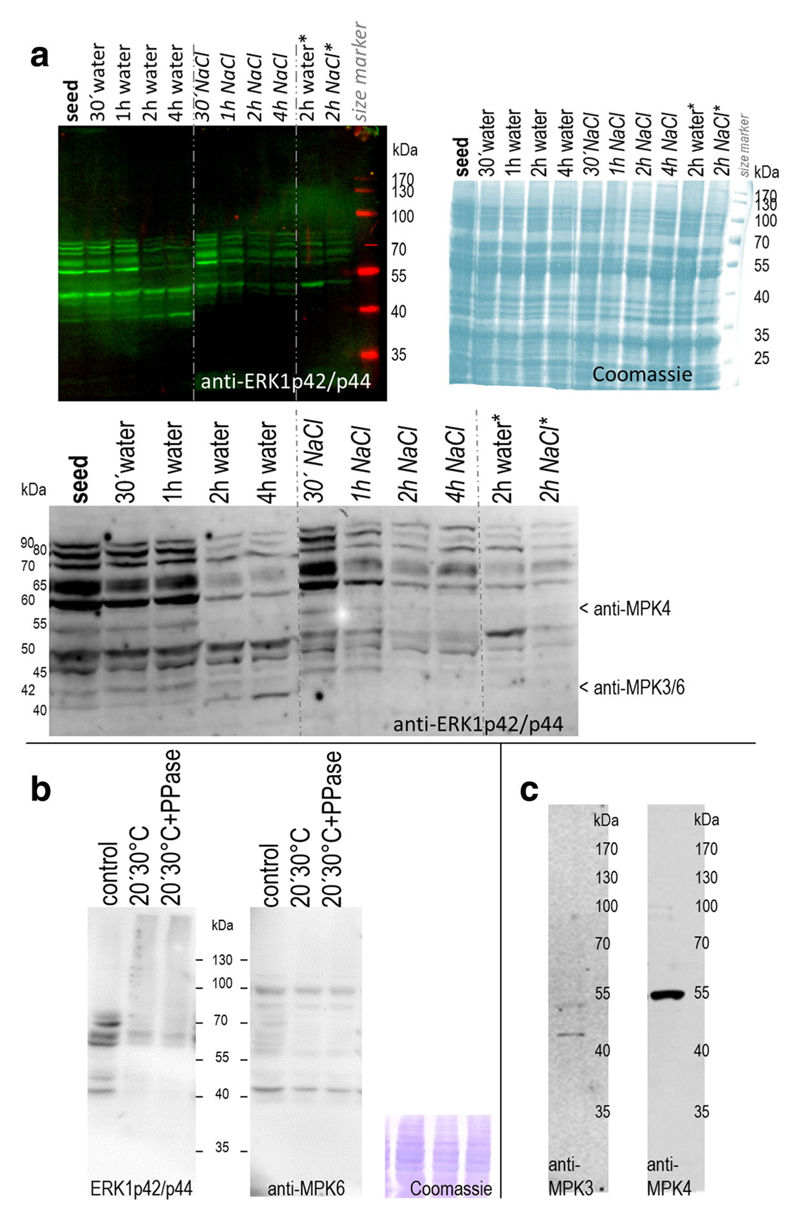
Fig. 1.
MAPKs in germinating and salinity-stressed seeds a MAPK activity profiles during germination and salt stress. Proteins (20 μg) were extracted from quinoa (Real_Bolivia) dry seeds and after imbibition in water or 400 mM NaCl. For the 2-h time point, extracts from an independent experiment were examined in parallel (columns at the right; marked with „*“). Active forms of MAPKs were detected by immunoblot analysis with anti-ERK1p42/p44 antibody and infrared dye-coupled secondary anti-body. The blot was subsequently dried to enhance signal intensity, and a magnification of the 35–100 kDa region is shown in the lower part of the image. Arrows indicate protein bands potentially corresponding to anti-AtMPK3, –MPK4, and -MPK6 hybridisation signals (based on data of Fig. 1b/c). Right: A duplicate gel, containing the same samples, was stained with Coomassie blue to document protein loading. b Specificity of ERK1p42/p44 antibody. Quinoa seed protein extracts were incubated on ice (control) or at 30 °C for 20 min, without or with added phosphatase. Active forms of MAPKs were detected by immunoblot analysis with anti-ERK1p42/p44 antibody (left). The same membrane was subsequently hybridised with anti-MPK6 antibody (right), followed by CBB staining (bottom). Comparison of the two blots suggests that anti-ERK1p42/p44 signal reduction in treated samples (lanes 2&3) arises from dephosphorylation, not from protein degradation. c Cross-reactivity test with anti-Arabidopsis MAPK antibodies. Antibodies directed against evolutionary conserved peptides in Arabidopsis MPK3 and MPK4 were used for immunoblot analysis of quinoa protein extracts from (15 min)-water-imbibed seeds; 20 μg loaded)