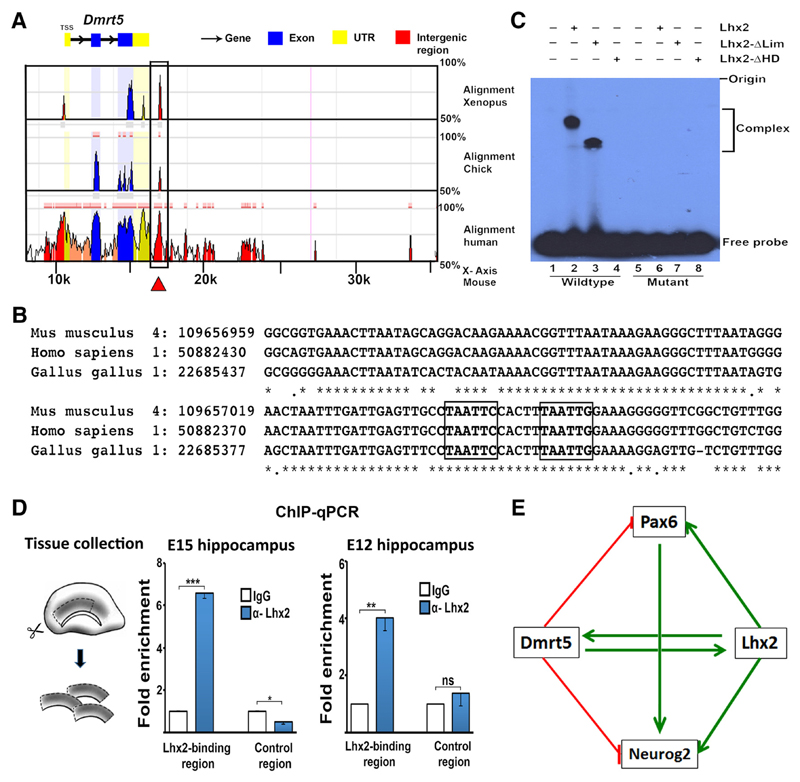
Figure 5. Lhx2 directly binds to the Dmrt5 locus within an evolutionarily conserved distal regulatory element.
A, B, ECR Browser view of the genomic region spanning 50 kb around the Dmrt5 locus. The Lhx2 binding region lies within a region in the 3′ UTR of Dmrt5 (A, red arrow) that is conserved across Xenopus, chick, and human. The Lhx2 binding site sequence reported in the literature (Folgueras et al., 2013) was identified in an element 6.4 kb downstream of Dmrt5. C, DNA EMSA was performed as mentioned in Materials and Methods. Radiolabeled oligonucleotides containing the wild-type sequence at the Lhx2 binding site and the putative binding site, or oligonucleotides mutated at both sites, were incubated with no protein (Lanes 1 and 5), or with in vitro synthesized full-length Lhx2 (Lane 2 and 6), or with Lhx2- Lim (Lanes 3 and 7), or with Lhx2- HD (Lanes 4 and 8). Incubation of the wild-type oligonucleotide with full-length Lhx2 (Lane 2) or with Lhx2-∆Lim (Lane 3) each resulted in retarded bands, whereas incubation with Lhx2-∆HD (Lane 4) or with no protein (Lane 1) did not lead to gel retardation. EMSA with the oligonucleotide containing the mutated sites yielded no gel retardation with any protein (Lanes 5– 8). D, Diagram illustrating the tissue collection from E15 and E12 embryonic hippocampi for ChIP followed by qPCR. In chromatin from both E15 and E12 tissue, Lhx2 displays significant enrichment at its binding site on the Dmrt5 locus compared with a control genomic region. E, A regulatory network of genetic interactions in the developing hippocampus. Lhx2 and Dmrt5 reciprocally regulate each other (Saulnier et al., 2013; De Clercq et al., 2016; this study). Lhx2 positively regulates Neurog2 and Pax6 (Shetty et al., 2013; this study), whereas Dmrt5 negatively regulates these factors (Saulnier et al., 2013). Neurog2 is also regulated by Pax6 (Scardigli et al., 2003). **p < 0.001, ***p < 0.0001.