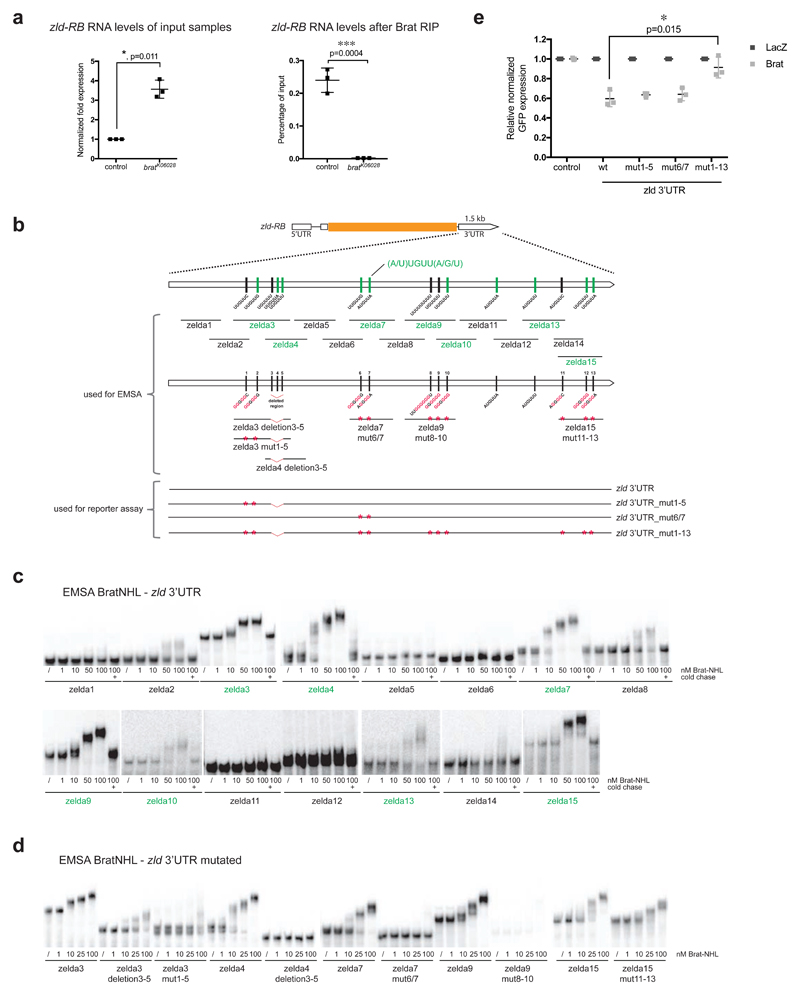
Figure 4. Brat directly targets the 3’UTR of zld via a specific binding motif.
(a) qPCR analysis of zld-RB transcript after Brat-RIP of control and bratK06028 mutant larval brain tissue. Left diagram represents zld-RB RNA levels of the Input. Right diagram represents zld-RB RNA levels after Brat-RIP experiment. (b) Schematic representation of zld-RB locus and the zld-RB 3’UTR fragments used in this study. Sites of the Brat-binding motif within the zld-RB 3’UTR are highlighted. Fragments, which bind to Brat-NHL are labeled in green; non-binding fragments in black. Nucleotide mutations and deletions are marked in red. (c, d) Recombinant Brat-NHL was incubated with 32P-labeled wild-type (c) or mutated (d) zld RNA fragments as indicated and analysed by native gel electrophoresis. Note that mutations of the Brat-binding sites in the zld-RB 3’UTR greatly impair RNA binding of Brat-NHL. (e) Drosophila S2 cells were cotransfected with GFP- zld-RB 3’UTR reporters as indicated together with full length Brat. RFP was used as a transfection control; LacZ was used as an overexpression control. Note that repression of GFP- zld-RB 3’UTR reporter bearing all Brat-binding site mutations is greatly reduced.
Pictures and blots are representative of three independent experiments.
Error bars represent standard deviation. Statistical analysis was done using T-test. **p<0.01.