Abstract
Purpose
Transcriptomic profiling of colorectal cancer (CRC) has led to identification of four consensus molecular subtypes (CMS1-4), which have prognostic value in stage II/III disease. More recently, the Colorectal Cancer Intrinsic Subtypes (CRIS) classification system has helped to define the biology specific to the epithelial component of colorectal tumors. However, the clinical value of these classifications in predicting response to standard-of-care adjuvant chemotherapy remains unknown.
Patients and Methods
Using samples from 4 European sites, we assembled a novel stage II/III CRC patient cohort and performed transcriptomic profiling on 156 samples, targeted sequencing and generated a tissue microarray to enable integrated “multi-omics” analyses. We also accessed data from 2 published stage II/III CRC patient cohorts: GSE39582 and GSE14333 (479 and 185 samples respectively)
Results
The epithelial-rich CMS2 subtype of CRC benefitted significantly from adjuvant chemotherapy treatment in both stage II and III disease (p=0.02 and p<0.0001 respectively), while the CMS3 subtype significantly benefitted in stage III only (p=0.00073). Following CRIS sub-stratification of CMS2, we observed that only the CRIS-C subtype significantly benefitted from adjuvant chemotherapy in stage II and III disease (p=0.0081 and p<0.0001 respectively), while CRIS-D significantly benefitted in stage III only (p=0.0034). We also observed that CRIS-C patients with low levels of CD8+ tumor-infiltrating lymphocytes were most at risk of relapse in both stage II and III disease (p=0.0031).
Conclusion
Patient stratification using a combination of transcriptional subtyping and CD8 immunohistochemistry analyses is capable of identifying poor prognostic stage II/III patients who benefit from adjuvant standard-of-care chemotherapy. These findings are particularly relevant for stage II disease, where the overall benefit of adjuvant chemotherapy is marginal.
Introduction
Colorectal cancer (CRC) has the 3rd highest worldwide incidence (~1.4 million new cases) and mortality rates (~780,000 deaths per year)1. Although 75% of patients with CRC present with operable disease (mainly stages II and III), around 40% of patients experience disease recurrence2. Importantly, compared to surgery alone, adjuvant chemotherapy improves survival in only ~3% of stage II patients; however, this rises to 15-20% for stage III. Overall, there is a clear need for treatment-stratifying biomarkers in stage II and III CRC and development of new therapeutic approaches for patients who do not benefit from standard 5-Fluorouracil (5FU)-based chemotherapy.
Significant advances have been made in molecular stratification of CRC, leading to the identification of four Consensus Molecular Subtypes (CMS1-4)3. Two of these subtypes map to previously identified pathological subtypes: CMS1 is enriched for microsatellite instability (MSI), is immune-rich and correlates with good prognosis; CMS4 is stromal-rich, with high levels of cancer-associated fibroblasts (CAFs) and has a relatively poor prognosis. CMS3 is defined by activation of multiple metabolic pathways, potentially due to its enrichment for KRAS mutations4. The epithelial-rich CMS2, termed the canonical subtype due to its robust upregulation of classic hallmarks of CRC (WNT and MYC pathways), is the largest group, accounting for ~40% of all tumors. Although CMS classification provides valuable prognostic information for early stage CRC, its usefulness for selecting patients for adjuvant chemotherapy is not clear5–7. Additionally, there are wide variations in clinical outcome within each CMS subtype, particularly CMS2; therefore, there is a clear need for refinement of this classification system.
In order to define the biology specifically driving neoplastic epithelial cells, the Colorectal Cancer Intrinsic Subtypes (CRIS) classification system was developed by transcriptional profiling of patient-derived xenografts (PDXs)6. Using this approach, five cancer epithelium-specific subtypes (CRISA-E) were identified. By focussing on tumor epithelium, the CRIS methodology can potentially identify aspects of neoplastic biology that would be masked by contributions from the tumor microenvironment (TME) when using the CMS approach8, 9. In this study, we used the epithelial-specific CRIS classification in combination with CMS to sub-stratify CMS2 tumors, in order to identify specific tumor-intrinsic traits associated with response to standard-of-care treatment.
Methods and Patient Cohorts. (Additional methods are provided in the Data Supplement)
Development of a multi-omics molecular pathology resource for CRC
Using a combination of transcriptome profiling, next generation sequencing (NGS) and tissue microarray generation, we have developed a unique resource for stage II and III CRC. This “Taxonomy” cohort was assembled from an initial cohort of 363 stage II and III patients from 4 European Centres (VHIO, Barcelona; St Vincent’s Dublin; University of Florence; and University of Aberdeen). This work was approved by the Medicine, Dentistry and Biomedical Sciences School Ethics Committee ref: 12/12v4. Within this cohort, samples from 194 patients with 50% or more tumor content as assessed by Hematoxylin and eosin (H&E) staining were subjected to RNA extraction, of these, 188 samples passed quality control (spectrophotometer A260/280: 1.68-2.08; 2 distinct peaks 18S and 28S on Bioanalyser). These 188 samples were subjected to RNA and DNA analysis.
Transcriptomics
High quality transcriptomics data were obtained for 156 stage II and III samples using the Almac Xcel™ array (Almac Diagnostics, Craigavon UK). All data analysis was performed using the R Statistical Package (version 3.4.1). All CEL files were loaded into R and processed using the makecdfenv, affy and limma packages. Residual technical batch effects were corrected using the Combat method within the sva package. All data have been deposited in the NCBI Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/geo/) (GSE103479). The clinical-pathological details of this cohort are provided in Table 1.
Table 1.
Table of clinical pathological details for the “Taxonomy” dataset and the GSE39582 stage II and II patient public dataset. Displayed are the numbers and percentages for each group. For age, mean lymph nodes examined and median follow-up (overall survival (OS)) the number, standard deviation and range are displayed. *5-FU, capecitabine or tomudex with oxaliplatin: 38; 5-FU or capecitabine: 27; 5-FU with irinotecan: 1. ** For the GSE14333 dataset the median follow-up measure was disease-free survival (DFS).
| Clinical variable | Taxonomy | GSE39582 (stage II/III) | GSE14333 (stage II/III) |
|---|---|---|---|
| Clinical Site (percent) | |||
| Aberdeen Barcelona Florence SVH Dublin |
27 (17.3) 63 (40.4) 17 (10.9) 49 (31.4) |
- | - |
| Mean age (sd, range), years | 69.37 (11.37, 37-94) | 67.67 (12.97, 22-97) | 65.70 (13.50, 26-92 |
| Sex (male/female) n (percent) | 88/68 (56.4/43.6) | 269/212 (56/44) | 98/87 (53/47) |
| TNM stage n (percent) | |||
| Stage II A Stage IIB Stage IIC Stage IIIA Stage IIIB Stage IIIC |
70 (44.9) 8 (5.1) 6 (3.8) 6 (3.9) 47 (30.1) 19 (12.2) |
Stage II 271 (56.3) Stage III 210 (43.7) |
Stage II 94 (50.8) Stage III 91 (49.2) |
| Treatment n (percent) | |||
| 5FU-based* Other No treatment Not recorded |
66 (42.31) - 87 (55.77) 3(1.92) |
149 (30.9) 61 (12.7) 269 (55.9) 2 (0.4) |
85 (46.0) - 100 (54.0) - |
| Mean Lymph nodes examined (sd, range) | 21.01 (13.84, 0-145) | - | - |
| Primary tumour localization (percent) | |||
| Left sided Right sided Not specified |
86 (55) 63 (40) 7 (5) |
282 (58.6) 199 (41.4) 0 |
77 (41.6) 85 (46.0) 23 (12.4) |
| Median follow-up (sd, range), months | 49.41 (39.67, 0-217.94) | 61.26 (38.83, 0-201) | 36.92 (26.05, 0.92-118.58) ** |
Data Analysis
The GSE3958210 and GSE14333311 CRC datasets were downloaded from the NCBI Gene Expression Omnibus (GEO) repository and their respective CEL files uploaded into R for data processing. The GSE39582 and GSE14333 datasets represent 585 fresh frozen (FF) and the dataset represents 290 FF surgically resected primary tumor samples respectively. Both datasets contain all stages (I-IV) and treated and untreated samples with accompanying clinical follow-up. Within the GSE39582 and GSE143333 datasets, only stage II and III patients were selected for further analysis (479 samples from GSE39582 and 185 samples from GSE14333). The clinical-pathological details of the GSE39582 and FSE14333 cohorts are provided in Table 1. Each dataset was subject to CMS and CRIS classification. The CMSclassifier R package was downloaded from github (https://github.com/Sage-Bionetworks/crcsc). The CRISclassifier R package was downloaded from Isella et al 6 and was implemented using the Nearest Template Prediction (NTP) method. Kaplan-Meier estimators and Cox proportional hazards regression analysis were assessed using the survival and survminer R packages. Correlations between CMS and CRIS subtypes were assessed using Caleydo plots, generated using Caleydo 3.1.5 software (www.caleydo.org).
TMA construction
In order to account for anticipated variations in protein expression and cellularity due to intra-tumoral heterogeneity, TMAs were generated with 9 cores per tumor, incorporating 3 cores each from central tumor (CT), invasive front (IF) and tumor-adjacent stroma-rich (SR) regions. In addition, where available, 3 cores of adjacent normal colonic tissue were arrayed.
Results
Molecular subgroups
We initially assessed the proportion of CMS and CRIS subtypes (Supplementary Table 1) 3, 6, 12, 13 present in our in-house multi-omics Taxonomy cohort (Table 1) and in 2 other independent publically-available cohorts, GSE39582 10 and GSE14333 11. These analyses revealed similar proportions of each CMS (Mann–Whitney paired t test, p value range:0.625-1) and CRIS subtype (Mann-Whitney paired t test, p value range: 0.8125-1) compared to the published proportions of CMS and CRIS subtypes (Supplementary Table 2) 3, 6.
Benefit from adjuvant 5FU-based chemotherapy in CMS
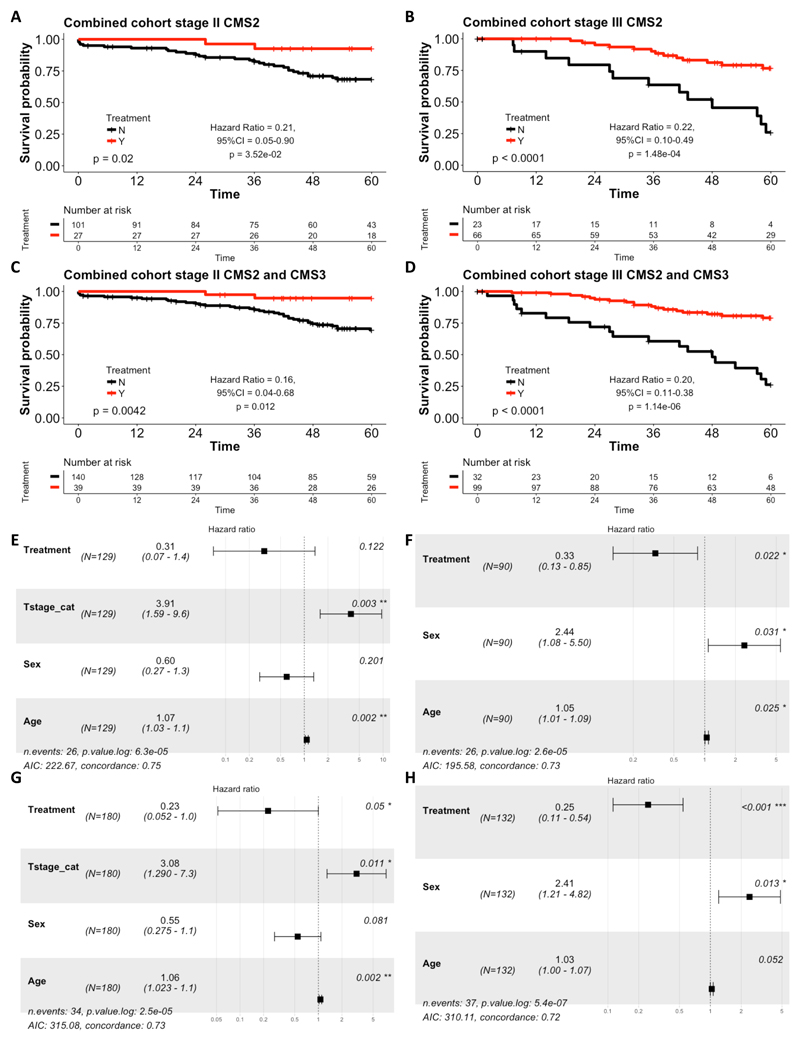
To determine benefit from adjuvant 5FU-based chemotherapy in CMS1-4, we used Kaplan-Meier analyses to assess overall survival (OS) of stage II and III patients, initially using the taxonomy cohort. Compared to patients treated by surgery-alone, there were non-significant trends particularly in stage III for CMS2 patients receiving chemotherapy to have improved OS (log-rank test p=0.13 for stage II and 0.056 for stage III; Supplementary Figures 1A and B). Similar results were obtained in the larger GSE39582 cohort for stage II (log-rank test p=0.071), while in stage III disease, this correlation reached significance (p=0.00075; Supplementary Figures 1C and D). When we combined the taxonomy and GSE39582 cohorts to increase statistical power, the benefit from adjuvant chemotherapy for CMS2 was significant in both stage II (log-rank test p=0.02, Hazard ratio for the chemotherapy-treated = 0.21 (Wald test p = 3.52x10-2); Figure 1A) and stage III (log-rank test p<0.0001, Hazard ratio for the chemotherapy-treated group = 0.22 (Wald test p = 1.48x10-4); Figure 1B). There was also significant benefit from adjuvant chemotherapy treatment in the stage III CMS3 subtype (log-rank test p=0.00073, Hazard ratio for the chemotherapy-treated group = 0.16 (Wald test p=2.95x10-3); Supplementary Figure 2D) and a trend for benefit from chemotherapy in the stage II CMS3 subtype; however, this failed to reach significance (log-rank test p=0.088; Supplementary Figure 2C). Notably, no significant benefit from adjuvant chemotherapy was observed in CMS1 or CMS4, although a non-significant trend was observed in CMS4 stage III (log-rank test p=0.089; Supplementary Figures 2A, B, E and F).
Figure 1.
Kaplan-Meier plots of 5 year overall survival (OS) for CMS2 patients who received adjuvant 5-FU-based treatment (red) and those who did not receive treatment (surgery alone, black), in (A) stage II combined (Taxonomy and GSE39582) cohort and (B) stage III combined (Taxonomy and GSE39582) cohort. Kaplan Meier plots for 5 year OS for combined CMS2 and CMS3 patients in: (C) stage II combined (Taxonomy and GSE39582) cohort; and (D) stage III combined (Taxonomy and GSE39582) cohort. Displayed is the log rank test, along with the Hazard ratio for the chemotherapy-treated group with 95% confidence intervals and Wald test of significance. Forest plots showing the results from the adjusted cox proportional hazards regression for: (E) stage II CMS2 combined cohort; (F) stage III CMS2 combined cohort; (G) stage II CMS2 and CMS3 combined cohort; and (H) stage III CMS2 and CMS3 combined cohort. The forest plots display the number of patients (N), the Hazard ratio for the chemotherapy-treated group with 95% confidence intervals and the Wald test of significance. In addition, the number of events and the log-likelihood ratio is also displayed. For the adjusted analyses, the data is stratified by treatment and adjusted for T stage, sex and age in stage II and for age and sex in stage III.
These results suggest that the more epithelial CMS2 and CMS3 subgroups benefit from adjuvant chemotherapy, whereas the CMS1 and CMS4 subgroups fail to benefit, particularly in the stage II setting. In support of this, in a combined analysis of the CMS2 and CMS3 subgroups, the benefit from adjuvant chemotherapy for the epithelial CMS2/3 was significant in both stage II (log-rank test p=0.0042, Hazard ratio for the chemotherapy-treated group = 0.16 (Wald test p = 0.012); Figure 1C) and stage III (log-rank test p<0.0001, Hazard ratio for the chemotherapy-treated group = 0.20 (Wald test p = 1.14x10-6); Figure 1D). In contrast, in a combined analysis of the CMS1 and CMS4 subgroups, no benefit from adjuvant chemotherapy was observed (Supplementary Figures 2G and H).
When the CMS2 subgroup results for stage II disease were adjusted for T stage (T4 versus T1-3), Age and Sex using Cox multiple regression, the significance of the benefit from chemotherapy was lost (Hazard ratio for the chemotherapy-treated group = 0.122, Wald test p=0.122, log likelihood ratio = 6.3x10-5; Figure 1E). However, in stage III disease, adjusting in this case for Age and Sex, the significance of benefit from adjuvant chemotherapy in CMS2 was maintained (Hazard ratio for the chemotherapy-treated group = 0.33, Wald test p=0.022, log likelihood p value = 2.6x10-5; Figure 1F). In Cox multiple regression analyses of the combined CMS2 and CMS3 epithelial subgroups, the significance of the benefit from chemotherapy was maintained in stage II (Hazard ratio for the chemotherapy-treated group = 0.23, Wald test p=0.05, log likelihood ratio = 2.5x10-5; Figure 1G) and stage III disease (Hazard ratio for the chemotherapy-treated group = 0.25, Wald test p<0.001, log likelihood p value = 5.4x10-7; Figure 1H).
Clinical implications of tumor-intrinsic stratification in CMS2
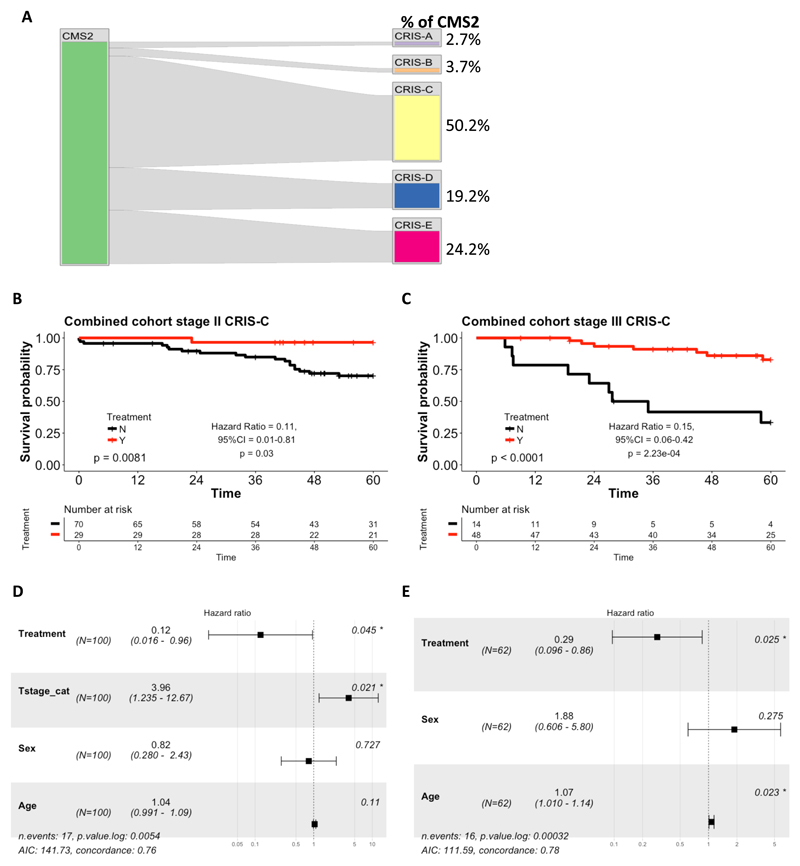
As previously reported6, there are limited associations between CMS and CRIS classification; for example, CMS4 is distributed relatively evenly between the 5 CRIS subtypes (Supplementary Figure 3). However, some clear patterns were observed: CMS3 predominantly maps to CRIS-A; CMS1 was distributed between CRIS-A and CRIS-B (Supplementary Figure 3). Importantly, CMS2 was almost exclusively distributed between CRIS-C, CRIS-D and CRIS-E (Figure 2A). Given that our data indicated that CMS2 tumors benefit from adjuvant standard-of-care chemotherapy and that this subtype is the largest numerically (Supplementary Table 2), we next investigated whether sub-stratification of CMS2 using the CRIS subtyping approach could identify a more specific subset of stage II/III patients who derive benefit from adjuvant chemotherapy. As the numbers of stage II and III patients in the individual cohorts are small when stratified into CRIS subgroups, we again combined the taxonomy and GSE39582 cohorts for these analyses. Of note, only the CRIS-C subgroup, which is the largest subgroup of CMS2 (50.2% (n=110), Figure 2A), displayed a significant benefit from adjuvant chemotherapy in both stage II (log-rank test p=0.0081, Hazard ratio for the chemotherapy-treated group = 0.12 (Wald test p = 0.03); Figure 2B) and stage III disease (log-rank test p<0.0001, Hazard ratio for the chemotherapy-treated group = 0.15 (Wald test p = 2.23x10-4; Figure 2C) or combined stage II/III (log-rank test p=3.5x10-4, Hazard ratio for the chemotherapy-treated group = 0.26 (Wald test p=8.8x10-4); Supplementary Figure 4A). These results were confirmed in a further independent cohort, GSE14333 (log-rank test p=0.02, Hazard ratio for the chemotherapy-treated group = 0.12 (Wald test p=0.05); Supplementary Figure 4B). In contrast, there was no significant benefit from adjuvant chemotherapy in stage II patients identified as CRIS-D (log-rank test p=0.28) or in stage II and III patients identified as CRIS-E (log-rank test p=0.37 and p=0.1 respectively); these analyses did however reveal significant benefit following adjuvant chemotherapy in stage III CRIS-D patients (log-rank test p=0.0034, Hazard ratio for the chemotherapy-treated group = 0.21 (Wald test p = 7.74x10-3); Supplementary Figures 5E-H). In the other 2 CRIS subgroups (CRIS-A and -B), no significant benefit from adjuvant chemotherapy was observed in either stage II or stage III disease, although a non-significant trend was observed in CRIS-A for stage III (p=0.057; Supplementary Figures 5A-B), consistent with this subgroup being enriched for CMS-3, where a significant benefit was observed (Supplementary Figures 2D).
Figure 2.
(A) Caleydo plots displaying the mapping of patient samples from the CMS2 subtype to the CRIS subtypes in the combined (Taxonomy and GSE39582) datasets. Plots were generated using Caleydo 3.1.5 software (www.caleydo.org). Kaplan-Meier plots of 5 year overall survival (OS) for: (B) stage II CRIS-C combined cohort; and (C) stage III CRIS-C combined cohort. Forest plots showing the results from the adjusted Cox proportional hazards regression for: (D) stage II; and (E) stage III CRIS-C in the combined cohort. The forest plots display the number of patients, the Hazard ratio for the chemotherapy-treated group with 95% confidence intervals and the Wald test of significance. In addition, the number of events and the log-likelihood ratio is also presented.
Importantly, when the CRIS-C results were adjusted for T Stage, Age and Sex using Cox multiple regression analyses, the hazard ratio for the chemotherapy-treated group associated with benefit from chemotherapy maintained significance in stage II disease (Hazard ratio for the chemotherapy-treated group = 0.12; Wald test p=0.045, log likelihood p value =0.0054; Figure 2D). Similarly, in stage III, adjusting for Age and Sex, the significant benefit from adjuvant chemotherapy was maintained (Hazard ratio for the chemotherapy-treated group = 0.29, Wald test p=0.025; log likelihood p value =3.2x10-4; Figure 2E), while for combined stage II and III disease, the Hazard ratio for the chemotherapy-treated group, adjusted for Stage, Sex and Age, was 0.20 (Wald test p<0.001, log likelihood p value =7.5x10-6; Supplementary Figure 4C). These results indicate that CRIS-C classification predicts benefit from adjuvant chemotherapy in both stage II and III patients independently of other clinicopathological factors.
Immunohistochemical assessment of CD8+ Tumor Infiltrating Lymphocytes
Although the benefit of adjuvant chemotherapy for CRIS-C patients is clear in stage II and stage III (Figure 2A-D), over 70% of stage II CRIS-C patients survive with surgery alone (Figure 2B). Thus, we sought to identify a routine method that could identify high-risk stage II CRIS-C patients who should be given adjuvant 5FU-based chemotherapy. Initially, we examined sequencing data from the Taxonomy and GSE39582 cohorts. As previously reported6, the CRIS-C subgroup is predominantly TP53 mutant and KRAS wild-type and almost exclusively BRAF wild-type (Supplementary Table 3). In line with this genotype, we also found that the CRIS-C subgroup is associated with left-sided tumours (Fishers exact test, CRIS-C against all others, p=6.27x10-10, 95% CI 2.26-5.48, odds ratio=3.48). Stratifying CRIS-C patients based on either TP53 or KRAS mutational status did not identify stage II (or stage III) CRIS-C patients at higher risk of relapse following surgery (data not shown).
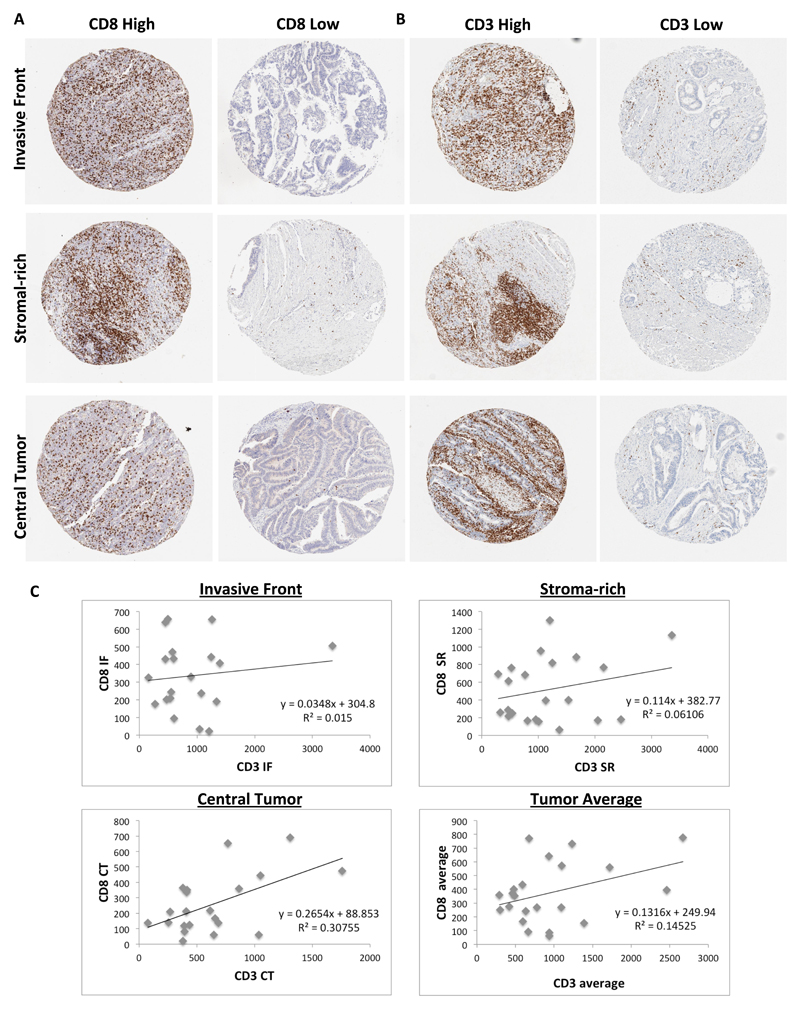
Given that stage II/III CRC patients with high levels of T cell infiltration have better prognoses than patients with low levels2, we next assessed whether T cell infiltration could be used to distinguish between low and high risk CRIS-C patients. In order to account for potential intra-tumoral heterogeneity (ITH) of putative biomarkers in specific tumor regions, TMAs were generated from the taxonomy cohort to incorporate 3 cores each from central tumor (CT), invasive front (IF) and tumor-adjacent stroma-rich (SR) regions. This unique design enables us to assess loco-regional variations in biomarker expression. CD8 and CD3 levels were defined using IHC, and patients were stratified into high and low groups using the median as the cut-off (representative CD8 and CD3 images, Figure 3A and B). Interestingly, the correlations between CD3 and CD8 scores were relatively low (Figure 3C).
Figure 3.
Representative images of samples displaying high and low CD8 (A) and CD3 (B) expression in each of the three tumor regions sampled: Invasive Front, Stromal-rich and Central Tumour. (C) Correlations between CD8 and CD3 at each of the three regions sampled: Invasive Front, Stromal-rich and Central Tumour and also the average expression. Displayed are the equations of the line and R2 value for the correlation.
In combined analyses of stage II/III disease, CRIS-C patients whose tumors had high levels of CD8-positive lymphocytes in the IF, SR and CT regions had a significantly better OS than those with low levels (log-rank test p=0.023, p=0.032 and p=0.011 respectively; Supplementary Figure 6A-C). We further assessed CD8-positive lymphocytes in histologically normal tissue adjacent to the tumor and found no difference in survival between high and low CD8 levels (log rank test p=0.72; Supplementary Figure 6D). Notably, no correlations were found between CD3-positive lymphocyte levels and prognosis (IF log-rank test p=0.55, SR log-rank test p=0.75, CT log-rank test p=0.8, normal log rank test p = 0.82; Supplementary Figure 7A-D).
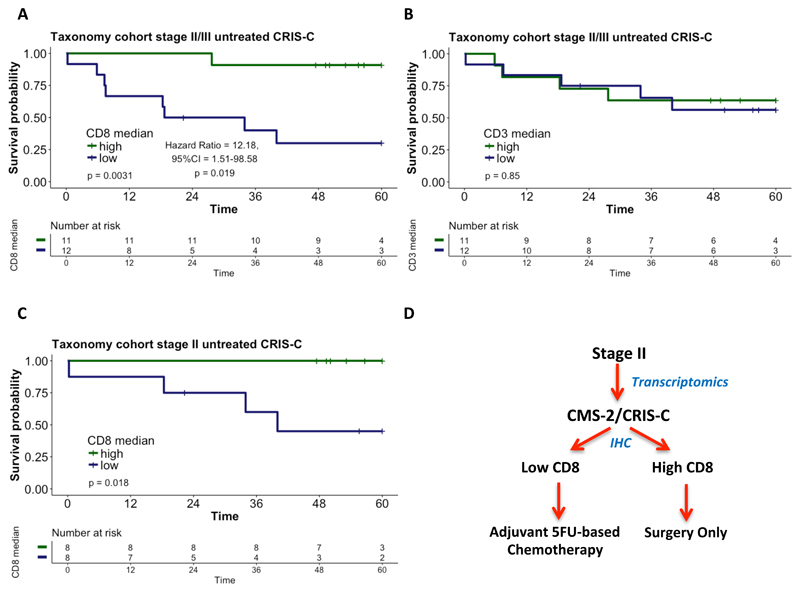
For each patient, the CD8 scores in each tumor sub-region correlated closely with one another (p<0.0001; Supplementary Table 4). We therefore combined the CD8 scores from each region where at least 2 out of 3 cores were present per region (CT, IF and SR) into an average score per patient. As expected based on the correlations for the individual regions (Supplementary Figure 6), applying this tumor average score to the combined stage II/III taxonomy cohort revealed that high levels of CD8-positive lymphocytes identified CRIS-C patients with good prognosis (log-rank p=0.0031, Hazard ratio for the CD8-low group = 12.18 (Wald test p = 0.0191); Figure 4A). In contrast, average CD3 scores, which again did not correlate closely with average CD8 scores (Figure 3C), were not prognostic (Figure 4B). Importantly, the tumor average CD8 score also robustly risk stratified stage II CRIS-C patients (log-rank p=0.018; Figure 4C). Notably, CD8 mRNA expression was unable to distinguish good and poor prognosis patients (log rank p=0.83; Supplementary Figure 7E and F), highlighting the need to use immunohistochemistry in combination with transcriptomic profiling for effective patient prognostication. Based on these findings, CRIS-C patients with high levels of CD8-positive lymphocytes have an excellent prognosis, suggesting that these patients do not require adjuvant chemotherapy. In contrast, as less than half of the patients with stage II CRIS-C/CD8 low tumors survive with surgery alone (Figure 4C) and given the significant benefit of 5FU-based chemotherapy in the CRIS-C subgroup (Figure 2), these data suggest that the CRIS-C/CD8 low subgroup within CMS2 should be treated with adjuvant chemotherapy (Figure 4D).
Figure 4.
Kaplan-Meier plots of: (A) CD8-high versus CD8-low stage II/III CRIS-C surgery-only, Taxonomy patients; (B) CD3-high versus CD3-low stage II/III CRIS-C surgery-only, Taxonomy patients; and (C) CD8-high versus CD8-low stage II CRIS-C surgery-only, Taxonomy patients. Patients were split into high (green) and low (blue) CD8 or CD3 groups using the median as the cut-off. Significance was assessed using a log rank test. (D) Diagram of a potential decision tree for treatment of stage II CRIS-C patients.
Discussion
Over a decade ago, the seminal paper from Galon et al. demonstrated the importance of anti-tumor immunity in CRC by showing the prognostic value of assessing immune infiltration14. More recently, based on gene expression patterns, it has been proposed that CRC is comprised of 4 distinct subtypes: the Consensus Molecular Subtypes (CMS)3; this classification incorporates gene expression profiles from tumor, stroma and immune cells. Very recently, an alternative transcriptomics-based classification has been proposed that focuses on gene expression exclusively within the tumor cell compartment; this CRIS classification approach maps closely to the tumor’s underlying mutations6 15.
In this study, we highlight the potential clinical utility of CMS for selecting stage III CRC patients for adjuvant chemotherapy, with a significant benefit from post-surgery chemotherapy observed in the epithelial-rich CMS2 and CMS3 subgroups (both alone and combined), but not in the more undifferentiated CMS1 or CMS4 subgroups16, 17. We show that further stratification of CMS2 tumors according to CRIS classification helps define the biological diversity that underpins the CMS2 molecular subtype, which is predominantly distributed between CRIS-C, CRIS-D and CRIS-E. Importantly, we show that this sub-stratification identifies CRIS-C and CRIS-D as the subgroups of patients within CMS2 that derive clear benefit from adjuvant chemotherapy in stage III disease.
In stage II CRC, the use of chemotherapy following surgery is still a matter of debate: therapeutic benefit from adjuvant chemotherapy is modest for this group as a whole, with an absolute improvement in survival of ~3%19–23. Currently, additional pathological characteristics such as obstruction, perforation, extramural venous invasion and T stage (T4) are used to identify poor prognostic stage II disease and guide the decision of whether or not to start chemotherapy treatment24. Additional methods of assessing risk of recurrence in the adjuvant disease setting have been the focus of many studies over recent years25–30, leading to the identification of the 12 gene Oncotype DX assay31. This Recurrence Score algorithm has been extensively clinically tested32–34 and been demonstrated to identify stage II patients who are at higher risk of recurrence and stage III patients who are at lower risk of recurrence35, 36. Additional signatures have been proposed, including the 18 gene prognostic classifier known as ColoPrint37 and the 634 gene prognostic classifier known as ColDX28. The present study indicates that the transcriptionally-definable CMS2/CRIS-C patient subgroup may be the cohort of patients within stage II disease that benefits from standard adjuvant 5FU-based chemotherapy. Importantly, this benefit was found to be independent of T stage. Moreover, none of the other CRIS subgroups derived significant benefit from adjuvant chemotherapy in the stage II setting.
Subsequently, we found that low levels of tumoral/peritumoral CD8+ lymphoid cells could identify CRIS-C stage II (and indeed stage III) patients most at risk of relapse following surgery and who therefore should be given adjuvant chemotherapy. These data correlate well with our previous study on the prognostic significance of immune-derived PD-L1 mRNA expression in CRC, in which we postulated that patients with low immune infiltrates would significantly benefit from adjuvant 5FU-based chemotherapy following surgery9, 38. The meta-analysis of the IDEA (International Duration Evaluation of Adjuvant chemotherapy) collaboration examined whether a 3-month duration of oxaliplatin-containing adjuvant chemotherapy (FOLFOX4, modified FOLFOX6 or XELOX) is as effective as a 6-month schedule in stage III CRC patients. This study found that the 3-month treatment was almost as effective as the 6-month treatment and reduced the risk of treatment-associated toxicity, concluding that a 3-month treatment would be more beneficial for “low-risk” (T1-3/N1 tumors) stage III patients18. Our study suggests that levels of CD8-positive lymphocytes could also be used to identify such low-risk patients, at least in the CRIS-C subgroup. Of note, CRIS-C is enriched for mutant TP53 and wild-type KRAS tumors6, but neither of these established molecular markers provided further information with regard to disease outcome within the CRIS-C subgroup.
Collectively, these results provide the first evidence of the predictive value of the now well-established CMS and more recently described CRIS transcription-based classification systems; they also emphasize the utility of combining CMS and CRIS subtyping in a sub-stratification strategy to maximize clinical benefit from adjuvant 5FU-based chemotherapy in stage II and III CRC. Additionally, CRIS classification, in combination with assessment of CD8 tumor infiltrating lymphocytes (TILs) would enable prospective identification of the CRIS-C/CD8-low stage II patients, who significantly benefit from adjuvant 5FU-based chemotherapy. We do however recognize that there are a number of limitations in the current study, which was conducted on a relatively small number of retrospective samples collected outside of clinical trials, and realise that this hypothesis-generating study now requires validation in either larger patient cohorts or stratified trial cohorts enriched for this patient subtype. Nonetheless, this study suggests that transcription-based classification systems such as CMS and CRIS have the potential to be developed into patient stratification tools and, when used alone or alongside other molecular pathology approaches (such as IHC), could enable selection of CRC patients most likely to benefit from adjuvant chemotherapy, whilst at the same time sparing non-responders the potentially harmful treatment-related adverse events and sequelae of chemotherapy. Importantly, the CRIS subtyping method uses gene expression from the tumour epithelial cells only and is independent of stromal-derived signals, therefore, the CRIS subgroups can be detected irrespective of the profiling technology used or the tissue source15; such robustness and reproducibility are critical for clinical translation. In conclusion, this study suggests that stage III CRIS-C and stage II CRIS-C/CD8 low patients would benefit from adjuvant 5-FU-based chemotherapy; this now requires further validation in larger patient cohorts.
Supplementary Material
Acknowledgments
This work was funded by Cancer Research UK: C212/A13721 and C11884/A24387
References
- 1.http://www.who.int/mediacentre/factsheets/fs297/en/ [Internet]. Available from: http://www.who.int/mediacentre/factsheets/fs297/en/
- 2.Van Cutsem E, Cervantes A, Adam R, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27:1386–1422. doi: 10.1093/annonc/mdw235. [DOI] [PubMed] [Google Scholar]
- 3.Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21:1350–1356. doi: 10.1038/nm.3967. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kerr EM, Gaude E, Turrell FK, et al. Mutant Kras copy number defines metabolic reprogramming and therapeutic susceptibilities. Nature. 2016;531:110–113. doi: 10.1038/nature16967. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lenz Heinz-Josef, Ou Fang-Shu, Venook Alan P, Hochster Howard S, Niedzwiecki Donna, Goldberg Richard M, et al. Mayer Robert J, Bertagnolli Monica M, Blanke Charles David, Zemla Tyler, Qu Xueping, et al. Impact of consensus molecular subtyping (CMS) on overall survival (OS) and progression free survival (PFS) in patients (pts) with metastatic colorectal cancer (mCRC): Analysis of CALGB/SWOG 80405 (Alliance) J Clin Oncol. 2017 May;35(15_suppl):3511–3511. 15_suppl:3511–3511, 2017. [Google Scholar]
- 6.Isella C, Brundu F, Bellomo SE, et al. Selective analysis of cancer-cell intrinsic transcriptional traits defines novel clinically relevant subtypes of colorectal cancer. Nat Commun. 2017;8:15107. doi: 10.1038/ncomms15107. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.http://www.motricolor.eu/project/
- 8.Morris JS, Kopetz S. Tumor microenvironment in gene signatures: Critical biology or confounding noise? Clin Cancer Res. 2016;22:3989–3991. doi: 10.1158/1078-0432.CCR-16-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunne PD, McArt DG, Bradley CA, et al. Challenging the cancer molecular stratification dogma: Intratumoral heterogeneity undermines consensus molecular subtypes and potential diagnostic value in colorectal cancer. Clin Cancer Res. 2016;22:4095–4104. doi: 10.1158/1078-0432.CCR-16-0032. [DOI] [PubMed] [Google Scholar]
- 10.Marisa L, de Reyniès A, Duval A, et al. Gene Expression Classification of Colon Cancer into Molecular Subtypes: Characterization, Validation, and Prognostic Value. PLoS Med. 2013 doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jorissen RN, Gibbs P, Christie M, et al. Metastasis-associated gene expression changes predict poor outcomes in patients with Dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15:7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calon A, Lonardo E, Berenguer-Llergo A, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet. 2015;47:320–329. doi: 10.1038/ng.3225. [Internet] [DOI] [PubMed] [Google Scholar]
- 13.Jass JR, Love SB, Northover JMA. A New Prognostic Classification of Rectal Cancer. Lancet. 1987;329:1303–1306. doi: 10.1016/s0140-6736(87)90552-6. [DOI] [PubMed] [Google Scholar]
- 14.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science (80- ) 2006;313:1960–4. doi: 10.1126/science.1129139. [Internet] [DOI] [PubMed] [Google Scholar]
- 15.Dunne PD, Alderdice M, O’Reilly PG, et al. Cancer-cell intrinsic gene expression signatures overcome intratumoural heterogeneity bias in colorectal cancer patient classification. Nat Commun. 2017;8:15657. doi: 10.1038/ncomms15657. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchins GGA, Treanor D, Wright A, et al. Intra-tumoural stromal morphometry predicts disease recurrence but not response to 5-fluorouracil - results from the QUASAR trial of colorectal cancer. Histopathology. 2017 doi: 10.1111/his.13326. [DOI] [PubMed] [Google Scholar]
- 17.Roepman P, Schlicker A, Tabernero J, et al. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer. 2013;134:552–562. doi: 10.1002/ijc.28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi Qian, Sobrero Alberto F, Shields Anthony Frank, Yoshino Takayuki, Paul James, Taieb Julien, Sougklakos Ioannis, Kerr Rachel, Labianca Roberto, Meyerhardt Jeffrey A, Bonnetain Franck, et al. Prospective pooled analysis of six phase III trials investigating duration of adjuvant (adjuv) oxaliplatin-based therapy (3 vs 6 months) for patients (pts) with stage III colon cancer (CC): The IDEA (International Duration Evaluation of Adjuvant chemother. J Clin Oncol. 2017;35 [Internet]. Available from: https://meetinglibrary.asco.org/record/147028/abstract. [Google Scholar]
- 19.Figueredo A, Charette ML, Maroun J, et al. Adjuvant therapy for stage II colon cancer: A systematic review from the cancer care Ontario program in evidence-based care’s gastrointestinal cancer disease site group. J Clin Oncol. 2004;22:3395–3407. doi: 10.1200/JCO.2004.03.087. [DOI] [PubMed] [Google Scholar]
- 20.Gray R, Barnwell J, McConkey C, et al. Adjuvant chemotherapy versus observation in patients with colorectal cancer: a randomised study. Lancet. 2007;370:2020–2029. doi: 10.1016/S0140-6736(07)61866-2. [Internet] [DOI] [PubMed] [Google Scholar]
- 21.Kuebler JP, Wieand HS, O’Connell MJ, et al. Oxaliplatin combined with weekly bolus fluorouracil and leucovorin as surgical adjuvant chemotherapy for stage II and III colon cancer: Results from NSABP C-07. J Clin Oncol. 2007;25:2198–2204. doi: 10.1200/JCO.2006.08.2974. [DOI] [PubMed] [Google Scholar]
- 22.Fang SH, Efron JE, Berho ME, W S. Dilemma of stage II colon cancer and decision making for adjuvant chemotherapy. J Am Coll Surg. 2014;219:1056–69. doi: 10.1016/j.jamcollsurg.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 23.McCleary NJ, Benson AB, 3rd, D R. Personalizing Adjuvant Therapy for Stage II/III Colorectal Cancer. Am Soc Clin Oncol Educ Book. 2017;37:232–245. doi: 10.1200/EDBK_175660. [DOI] [PubMed] [Google Scholar]
- 24.Benson AB, Schrag D, Somerfield MR, et al. American Society of Clinical Oncology recommendations on adjuvant chemotherapy for stage II colon cancer. J Clin Oncol. 2004;22:3408–19. doi: 10.1200/JCO.2004.05.063. [Internet] [DOI] [PubMed] [Google Scholar]
- 25.Maak M, Simon I, Nitsche U, et al. Independent Validation of a Prognostic Genomic Signature (ColoPrint) for Patients With Stage II Colon Cancer. Ann Surg. 2013;257:1053–1058. doi: 10.1097/SLA.0b013e31827c1180. [Internet]. Available from: http://content.wkhealth.com/linkback/openurl?sid=WKPTLP:landingpage&an=00000658-201306000-00011. [DOI] [PubMed] [Google Scholar]
- 26.Kelley RK, Venook AP. Prognostic and predictive markers in stage II colon cancer: Is there a role for gene expression profiling? Clin Colorectal Cancer. 2011;10:73–80. doi: 10.1016/j.clcc.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Y, Casey G, Lavery IC, et al. Development of a Clinically Feasible Molecular Assay to Predict Recurrence of Stage II Colon Cancer. J Mol Diagnostics. 2008;10:346–354. doi: 10.2353/jmoldx.2008.080011. [Internet]. Available from: http://linkinghub.elsevier.com/retrieve/pii/S1525157810601717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennedy RD, Bylesjo M, Kerr P, et al. Development and independent validation of a prognostic assay for stage II colon cancer using formalin-fixed paraffin-embedded tissue. J Clin Oncol. 2011;29:4620–4626. doi: 10.1200/JCO.2011.35.4498. [Internet] [DOI] [PubMed] [Google Scholar]
- 29.Barrier A, Boelle P-Y, Roser F, et al. Stage II colon cancer prognosis prediction by tumor gene expression profiling. J Clin Oncol. 2006;24:4685–4691. doi: 10.1200/JCO.2005.05.0229. [DOI] [PubMed] [Google Scholar]
- 30.T J, S R, R P, et al. Additional validation of a genomic signature (ColoPrint) for the risk stratification of stage II colon cancer patients. Ann Oncol. 2010;21:vi17. [Internet]. Available from: http://ovidsp.ovid.com/ovidweb.cgi?T=JS&PAGE=reference&D=emed9&NEWS=N&AN=70186937. [Google Scholar]
- 31.O’Connell MJ, Lavery I, Yothers G, et al. Relationship between tumor gene expression and recurrence in four independent studies of patients with stage II/III colon cancer treated with surgery alone or surgery plus adjuvant fluorouracil plus leucovorin. J Clin Oncol. 2010;28:3937–3944. doi: 10.1200/JCO.2010.28.9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray RG, Quirke P, Handley K, et al. Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol. 2011;29:4611–9. doi: 10.1200/JCO.2010.32.8732. [Internet] [DOI] [PubMed] [Google Scholar]
- 33.Venook AP, Niedzwiecki D, Lopatin M, et al. Biologic determinants of tumor recurrence in stage II colon cancer: Validation study of the 12-gene recurrence score in cancer and leukemia group B (CALGB) 9581. J Clin Oncol. 2013;31:1775–1781. doi: 10.1200/JCO.2012.45.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yothers G, O’Connell MJ, Lee M, et al. Validation of the 12-gene colon cancer Recurrence Score in NSABP C-07 as a predictor of recurrence in patients with stage II and III colon cancer treated with fluorouracil and leucovorin (FU/LV) and FU/LV plus oxaliplatin. J Clin Oncol. 2013;31:4512–4519. doi: 10.1200/JCO.2012.47.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cartwright T, Chao C, Lee M, et al. Effect of the 12-gene colon cancer assay results on adjuvant treatment recommendations in patients with stage II colon cancer. Curr Med Res Opin. 2014;30:321–328. doi: 10.1185/03007995.2013.855183. [Internet] [DOI] [PubMed] [Google Scholar]
- 36.Srivastava G, Renfro LA, Behrens RJ, et al. Prospective multicenter study of the impact of oncotype DX colon cancer assay results on treatment recommendations in stage II colon cancer patients. Oncologist. 2014;19:492–497. doi: 10.1634/theoncologist.2013-0401. [Internet]. Available from: http://theoncologist.alphamedpress.org/content/19/5/492.full.pdf#page=1&zoom=auto,-16,783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salazar R, Roepman P, Capella G, et al. Gene expression signature to improve prognosis prediction of stage II and III colorectal cancer. J Clin Oncol. 2011;29:17–24. doi: 10.1200/JCO.2010.30.1077. [DOI] [PubMed] [Google Scholar]
- 38.Dunne PD, McArt DG, O’Reilly PG, et al. Immune-Derived PD-L1 Gene Expression Defines a Subgroup of Stage II/III Colorectal Cancer Patients with Favorable Prognosis Who May Be Harmed by Adjuvant Chemotherapy. Cancer Immunol Res. 2016;4:582–91. doi: 10.1158/2326-6066. [Internet] [DOI] [PubMed] [Google Scholar]
- 39.https://gatkforums.broadinstitute.org/gatk/discussion/3060/how-should-i-pre-process-data-from-multiplexed-sequencing-and-multi-library-designs
- 40.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [Internet] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bankhead P, Loughrey MB, Fernández JA, et al. QuPath : Open source software for digital pathology image analysis. bioRxiv. 2017:1–27. doi: 10.1038/s41598-017-17204-5. [Internet]. Available from: http://biorxiv.org/content/early/2017/03/06/099796?rss=1&utm_source=dlvr.it&utm_medium=twitter. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.