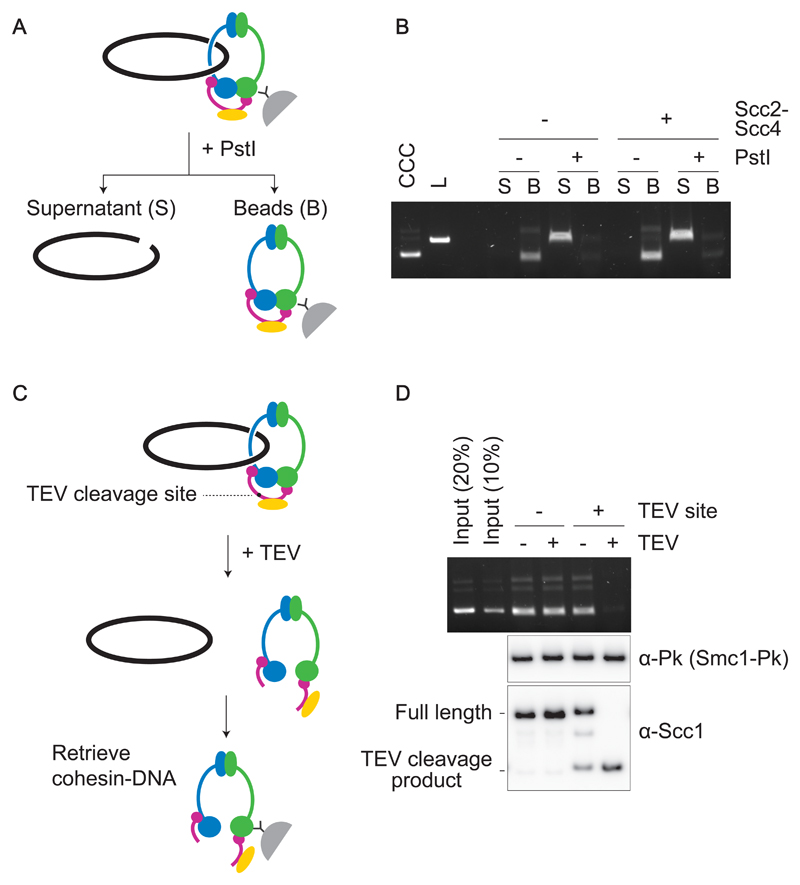
Figure 3. Topological DNA embrace by the budding yeast cohesin ring.
(A) Schematic of DNA-release by DNA linearization. Immobilized cohesin-DNA complexes were incubated in the presence or absence of PstI. The supernatant fraction (S) as well as beads-bound fraction (B) were collected, and DNA in each fraction was analyzed by agarose gel electrophoresis. (B) Gel image of an experiment as outlined in (A). Cohesin loading was performed with or without Scc2-Scc4. Covalently Closed Circular (CCC) and Linear (L) forms of the input DNA were included as a comparison. (C) Schematic of DNA-release by cohesin cleavage. (D) Wild type and TEV protease (TEV)-cleavable cohesin were loaded onto DNA, then TEV protease was added to half of the reaction. Cohesin was retrieved and recovered DNA analyzed by agarose gel electrophoresis. Scc1 cleavage was monitored by immunoblotting. Note that TEV-cleavable cohesin was partially cleaved even without TEV addition. This could be due to similarities between the TEV and PreScission protease recognitions sites, the latter was used during the cohesin purification.