Abstract
Hypoxia plays a critical role at cellular and physiological levels in all animals. The responses to chronic hypoxia are, at least substantially, orchestrated by activation of the hypoxia inducible transcription factors (HIFs), whose stability and subsequent transcriptional activation are regulated the by HIF hydroxylases. Factor inhibiting HIF (FIH), initially isolated as a HIFα interacting protein following a yeast two-hybrid screen, is an asparaginyl hydroxylase that negatively regulates transcriptional activation by HIF. This study aimed to define mechanisms that govern transitions of FIH between nucleus and the cytoplasm. We report that FIH accumulates in the nucleus within a short time window upon hypoxia treatment. We provide evidence, based on the application of genetic interventions and small molecule inhibition of the HIF hydroxylases, that the nuclear localization of FIH is governed by two opposing processes: nuclear entry by “coupling” with HIF1α for importin β1-mediated nuclear import and active export via a Leptomycin B-sensitive exportin1-dependent pathway.
Keywords: 2-oxoglutarate (2-OG), dioxygenase inhibitors, FIH (factor inhibiting HIF), HIF asparaginyl hydroxylase, hypoxia, nuclear translocation
Introduction
As solid tumors grow and oxygen becomes limiting, hypoxia triggers cellular and physiological events (Ratcliffe, 2013). The hypoxia-inducible factors (HIFs) are upregulated upon hypoxia and are key factors in coordinating cellular responses to hypoxia. HIF is an α, β – heterodimer that binds DNA at hypoxia response elements (HREs) containing a core “RCGTG” sequence (Kaelin and Ratcliffe, 2008). There are three HIFα proteins in higher metazoans, with HIF1α and HIF2α being the most extensively studied. HIF1α and HIF2α are closely related, and both activate HRE-dependent gene transcription. Nevertheless, HIF1α and HIF2α play non-redundant roles with distinct transcriptional targets (Kaelin and Ratcliffe, 2008; Ratcliffe, 2007). Levels of HIFα, but not HIFβ, are strongly regulated by oxygen availability, as is the transcriptional activity of HIF.
As a key regulator of the response of mammalian cells to oxygen deprivation and a critical player in the adaptation of tumor cells to a hypoxic microenvironment, the regulation of the stability and subsequent trans-activational function of HIFα is of major biomedical importance. Under well-oxygenated conditions, HIFα is hydroxylated at prolyl residues by members of the prolyl hydroxylase domain (PHD) family (Myllyharju, 2013). Hydroxylation of these prolyl residues generates a binding site for the von Hippel-Lindau (pVHL) tumor suppressor protein, which is a component of a ubiquitin E3 ligase complex. As a result, HIFα is polyubiquitylated and subjected to proteasomal degradation when oxygen is available. The PHD proteins belong to the Fe (II) and 2-oxoglutarate (2-OG)-dependent oxygenase superfamily, whose activity is dependent on oxygen. The kinetic properties of the PHDs enable the rate of HIF hydroxylation in cells to be suppressed by hypoxia. Under low oxygen conditions, or in cells lacking functional pVHL, HIFα accumulates, dimerizes with HIFβ, translocates to the nucleus, and transcriptionally activates multiple genes, including genes involved in erythropoiesis, angiogenesis, autophagy, and energy metabolism (Kaelin and Ratcliffe, 2008).
Factor inhibiting HIF (FIH), another Fe (II)- and 2-OG-dependent dioxygenase, hydroxylates a conserved asparagine residue within the HIFα C-terminal activation domain (CAD), a post-translational modification that blocks interactions between the HIFα CAD and the transcriptional activator/histone acetyl transferences CBP/p300 (Elkins et al., 2003; Hewitson et al., 2002; Lando et al., 2002a; Lando et al., 2002b; Mahon et al., 2001; McNeill et al., 2002). FIH has multiple other substrates, including members of the ankryin repeat domain (ARD) protein family (Cockman et al., 2006; Cockman et al., 2009; Coleman et al., 2007; Janke et al., 2013; Karttunen et al., 2015; Zheng et al., 2008). Because HIFα, ARD-containing proteins and other substrates can be located in different cellular compartments, processes that affect the subcellular location of FIH will influence its substrate selection and subsequently, its biological functions, including the regulation of metabolism (Peng et al., 2012a; Scholz et al., 2016; Sim et al., 2018; Zhang et al., 2010), keratinocyte differentiation (Peng et al., 2012b), vascular endothelial cell survival (Kiriakidis et al., 2015), tumour growth (Kuzmanov et al., 2012; Pelletier et al., 2012) and metastasis (Kang et al., 2017) as well as Wnt signalling (Rodriguez et al., 2016).
FIH is ubiquitously expressed in most types of cultured cells and human tissues (Bracken et al., 2006; Stolze et al., 2004). In live cells, overexpressed eGFP-tagged FIH is primarily observed in the cytoplasm, with a low level of nuclear protein (Metzen et al., 2003). Consistent with this observation, immunofluorescence analyses of endogenous FIH in cultured HEK 293T cells detected FIH protein predominantly in the cytoplasm (Linke et al., 2004; Stolze et al., 2004). A wide range of human tissues analysed by immunofluorescence also manifests mostly cytoplasmic staining, but for cell types expressing notably high levels of endogenous FIH, nuclear staining was also observed (Soilleux et al., 2005). In clinicopathological studies of human cancer, nuclear localization of FIH was reported to be a positive factor associated with good prognosis. This observation was independent of other more conventional features, including histopathological grading, tumour size, and spread to lymph nodes (Kroeze et al., 2010; Tan et al., 2007). Understanding the factors regulating FIH localization is therefore of both biological and medical interest. A previous study reported that hypoxia induces nuclear FIH (Liang et al., 2015), but the underlying mechanism remains unknown. Here we report a detailed time course analysis on the effects of hypoxia and HIF hydroxylase inhibition on FIH localization. The results reveal that FIH accumulates in the nucleus upon exposure to hypoxia within a short timeframe, and FIH enters and exits the nucleus via HIF1α/importin β1- and Leptomycin B-sensitive exportin1 (CRM1)-dependent pathways, respectively.
Results
Hypoxia induces nuclear entry of FIH
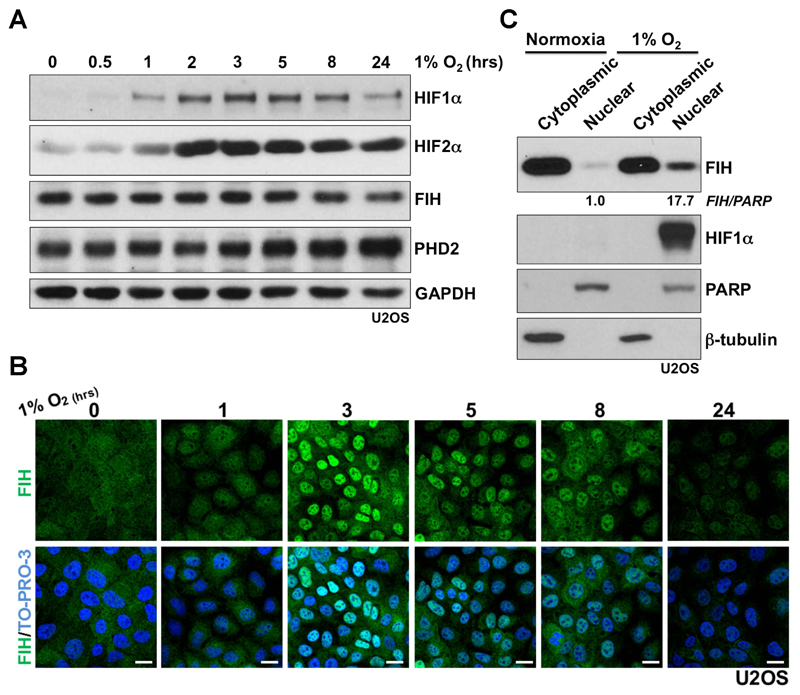
To investigate whether FIH nuclear import is regulated by hypoxia, a detailed time course of hypoxia (1% atmospheric O2) treatment was performed using human osteosarcoma U2OS cells. As shown in Fig. 1A, an overall increase in HIF1α protein level manifests after a 1-hour hypoxia incubation, with maximal induction being observed by western blots around 3 hours under hypoxia. After 8 hours under hypoxia, an apparent decrease in the HIF1α level was observed. The latter may due to the up-regulation of prolyl hydroxylase domain-containing protein PHD2, or other PHD isoforms, which are HIF target genes (Epstein et al., 2001; Marxsen et al., 2004); PHD2 is a major regulator of HIF1α steady state levels in many cells (Berra et al., 2003; Epstein et al., 2001). Unlike HIF1α, an increase in HIF2α protein level was observed and sustained under hypoxia for at least 24 hours. The FIH total protein level was not altered upon hypoxia within the limits of detection. However, immunofluorescence studies revealed a striking change in FIH localization on hypoxia treatment – clear nuclear accumulation of FIH was observed 3 hours post hypoxia treatment (Fig. 1B). FIH remained localized in the nucleus for several hours, but nuclear FIH was greatly reduced after 24 hours (Fig. 1B). The FIH immunofluorescence staining is specific since no signal was detected in FIH siRNA-transfected U2OS cells either in normoxia or hypoxia (Supplementary Fig. 1A). Similar results concerning the effects of hypoxia on FIH localization were obtained with both the human colon cancer cells HKe3 (Wang et al., 2014; Wang et al., 2010) and human breast cancer cells MCF-7 (Supplementary Fig. 1B and C). The hypoxia-induced nuclear accumulation of FIH observed by immunofluorescence staining was further supported by a subcellular fractionation assay that detected a > 17 fold increase in FIH protein in the nuclear fractions in U2OS cells upon hypoxia treatment (Fig. 1C). HIF1α was only observed to be present in the nuclear fraction upon hypoxia.
Figure 1. Evidence that hypoxia induces nuclear entry of FIH.
(A) Protein levels of HIF1α, HIF2α, FIH and PHD2 in U2OS cells upon hypoxia (1% O2) treatment at the indicated time points. GAPDH was used as a loading control.
(B) Immunofluorescence staining of FIH (green) in U2OS cells upon hypoxia (1% O2) at the indicated time points. TO-PRO-3 (blue) was used to stain nuclei. Scale bar: 20 µm.
(C) Protein levels of FIH and HIF1α from cytoplasmic or nuclear fractions in U2OS cells in normoxia or hypoxia (1% O2, 3 hours). β-tubulin and PARP were used as loading controls for the cytoplasmic and nuclear fractions respectively. Figures beneath lanes 2 and 4 are relative intensities of nuclear FIH in normoxia and hypoxia. Note that different quantity of cytoplasmic versus nuclear extract were loaded.
Nuclear entry of FIH is mainly HIF1α-dependent, and requires inhibition of FIH enzyme activity
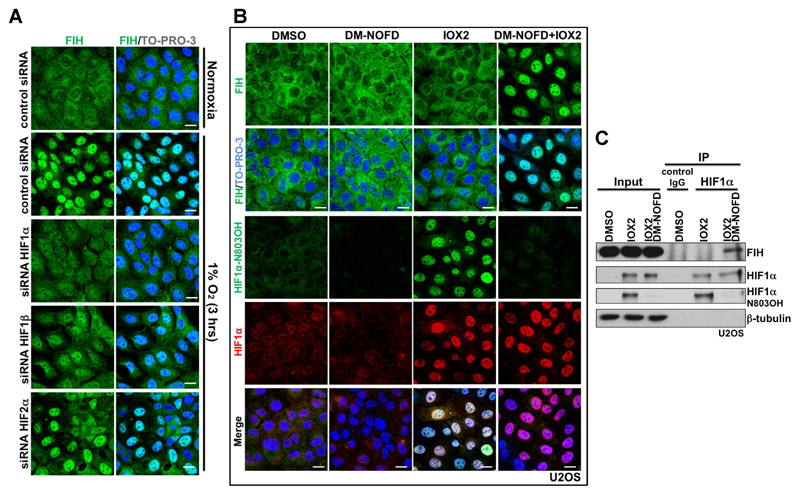
It was reported that in breast cancer, nuclear FIH expression showed a significant positive correlation with nuclear HIF1α expression (Tan et al., 2007). In addition, in the current work we observed that the time course of induction of HIF1α upon hypoxia correlates with that of nuclear accumulation of FIH. We thus hypothesized that nuclear entry of FIH is HIF1α-dependent. To test this, we depleted HIF1α, HIF2α or HIF1β using siRNAs in U2OS cells then exposed the cells to hypoxia (Supplementary Fig. 2A). HIF1α depletion abolished hypoxia-induced nuclear FIH (Fig. 2A, Supplementary Fig. 2B). HIF1β depletion also affected hypoxia-induced nuclear FIH, but to a lesser extent than HIF1α (Fig. 2A, Supplementary Fig. 2B), possibly via down-regulation of HIF1α (Supplementary Fig. 2A) (Chilov et al., 1999). Although the average intensity of nuclear FIH was decreased following HIF2α depletion, the percentage of nuclear FIH positive cells following HIF2α depletion was not significantly changed (Fig. 2A, Supplementary Fig. 2B).
Figure 2. Nuclear entry of FIH is mainly HIF1α-dependent, and requires inhibition of FIH enzymatic activity.
(A) Immunofluorescence staining of FIH (green) in U2OS cells with indicated treatments. U2OS cells were transfected with the indicated siRNA for 3 days, followed by culture in normoxia (20 % O2) or 3 hours’ hypoxia (1 % O2). TO-PRO-3 (blue) was used to stain nuclei. Scale bar: 20 µm.
(B) Immunofluorescence staining of FIH (green), HIF1α N803OH (green) and HIF1α (red) in U2OS cells treated with DMSO, DM-NOFD (1 mM), IOX2 (0.25 mM), or DM-NOFD (1 mM) plus IOX2 (0.25 mM) for 3 hours. TO-PRO-3 (blue) was used to stain nuclei. Scale bar: 20 µm.
(C) Protein levels of FIH, HIF1α and HIF1α N803OH in U2OS cells with indicated treatments. β-tubulin was used as a loading control. Total cell lysates from U2OS cells with indicated treatments were immunoprecipitated with an anti-HIF1α antibody.
To further define the dependence of FIH nuclear localization on the stabilization of HIF1α, we exposed cells to small molecule inhibitors of the HIF hydroxylases with differential selectivity against FIH and the PHDs. To monitor the action of these compounds in cells under the conditions of our experiments, we deployed an antibody which specifically recognizes N803 hydroxylated HIF1α, as assessed by mass spectrometry and reactivity against synthetic peptides (Lee et al., 2008; Tian et al., 2011). This demonstrated that, as expected, these compounds displayed differential activity against FIH and induced HIF1α in a form that is (IOX2, FG2216, DFO) or is not (DMOG, IOX1, VGB10B/IOX4) hydroxylated on HIF1α N803 (Supplementary Fig. 2C). Immunostaining of aliquots of the same cells for FIH revealed a striking correlation between the nuclear localization of FIH and those conditions in which HIF1α was induced in a form where N803 hydroxylation was undetectable (Supplementary Fig. 2D). These results indicated that the hydroxylation status of HIF1α at N803 was affecting the nuclear localization of FIH. Although HIF1α that was not hydroxylated on N803 could not be measured directly, the findings suggested that it was the nuclear accumulation of non-hydroxylated HIF1α that was responsible for FIH relocation. We postulated that this was a result of increased binding of FIH to this unhydroxylated HIF1α, as would be expected from previous work demonstrating that catalytic inhibitors of FIH promote binding to its substrates (Cockman et al., 2009). To test this more directly, we exposed cells to specific inhibitors of both the PHDs (IOX2) (Chan et al., 2016), and FIH (DM-NOFD) (McDonough et al., 2005), or both compounds in combination (Fig. 2B and C). As expected, IOX2 induced HIF1α in a form that was substantially hydroxylated on N803, whereas additional exposure to DM-NOFD suppressed the N803 hydroxylation (Fig. 2B and C). Under these conditions, clear nuclear localization of FIH was observed when IOX2 was combined with DM-NOFD but not when it was used alone (Fig. 2B). Consistent with the hypothesis that this reflected binding of FIH to unhydroxylated HIF1α, immunoprecipitation revealed binding of FIH to HIF1α was induced when N803 hydroxylation of HIF1α was suppressed by DM-NOFD (Fig. 2C).
Taken together these findings suggest that FIH enters the nucleus in association with its substrate, HIF1α, and that this process is enhanced by catalytic inhibition.
FIH enters and exits the nucleus via HIF1α/importin β1- and Leptomycin B-sensitive exportin1 (CRM1)-dependent pathways, respectively
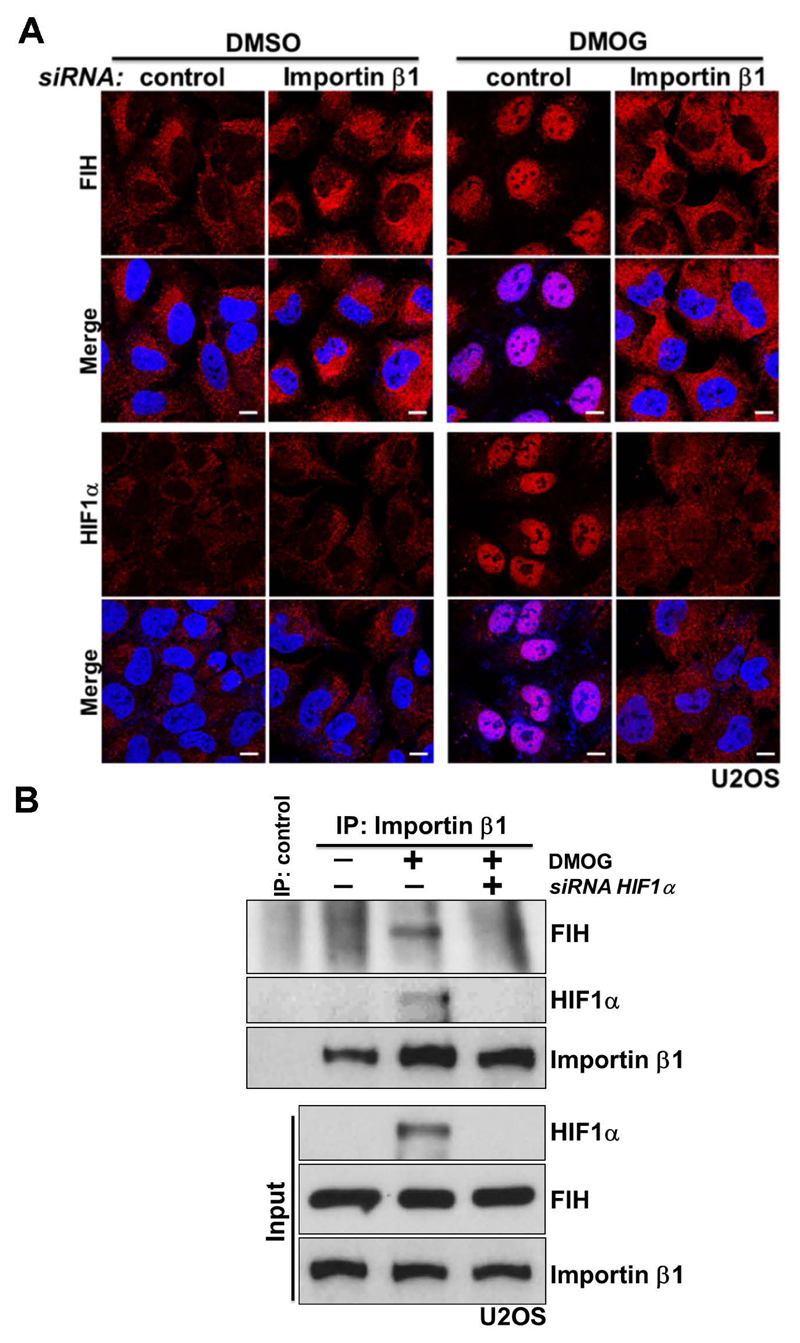
FIH has 349 residues and forms a ~80 kDa homodimer in solution (Dann et al., 2002; Elkins et al., 2003), which is essential for its efficient catalysis (Lancaster et al., 2004). The transport of proteins larger than ~40 kDa between the nucleus and cytoplasm through the nuclear pore complex (NPC) is a spatially and temporally controlled process (Adams and Wente, 2013; Aitchison and Rout, 2012). For nuclear import, target proteins using the classical nuclear import system bind to dimeric complexes of importin α/β proteins (Tran et al., 2014). Unpublished data from Kosyna et al. indicate that FIH is not interacting with importins α and β (Depping et al., 2015). The observations suggest that the nuclear import of FIH likely involves other proteins, consistent with the hypothesis that its nuclear entry is mediated, at least under these conditions, by association with HIF1α. A classical importin α/β-dependent bipartite NLS is present at the C-terminus of human HIFα (Depping et al., 2008). As anticipated, siRNA-dependent depletion of importin β1 blocked the nuclear accumulation of HIF1α induced by DMOG treatment (Supplementary Fig. 3; Fig. 3A, Row - HIF1α). A lower level of HIF1α protein was detected in DMOG-treated importin β1-depleted cells compared to DMOG-treated control cells, suggesting that the nuclear retention of HIF1α somehow reduces its degradation (Supplementary Fig. 3). Importantly, under the same conditions, DMOG-induced nuclear entry of FIH was abolished upon importin β1 depletion (Fig. 3A, Row - FIH). Consistent with this observation, DMOG treatment induced the association of importin β1 with HIF1α and FIH; while the binding between FIH and importin β1 was HIF1α-dependent, since knockdown of HIF1α abolished the FIH-importin β1 interaction (Fig. 3B).
Figure 3. FIH complexes with importin β1 via HIF1α for nuclear import.
(A) Immunofluorescence staining of FIH (red) or HIF1α (red) in U2OS cells with indicated treatments. U2OS cells were transfected with control siRNA or Importin β1 siRNA for 3 days, followed by treatment with DMSO or DMOG (1 mM) for 3 hours. DAPI (blue) was used to stain nuclei. Scale bar: 10 µm.
(B)Total cell lysates from U2OS cells with indicated treatments were immunoprecipitated with an anti-importin β1 antibody or control IgG. FIH, HIF1α and importin β levels are indicated. U2OS cells were transfected with control siRNA or HIF1α siRNA for 3 days, followed by treatment with DMSO or DMOG (1 mM) for 3 hours.
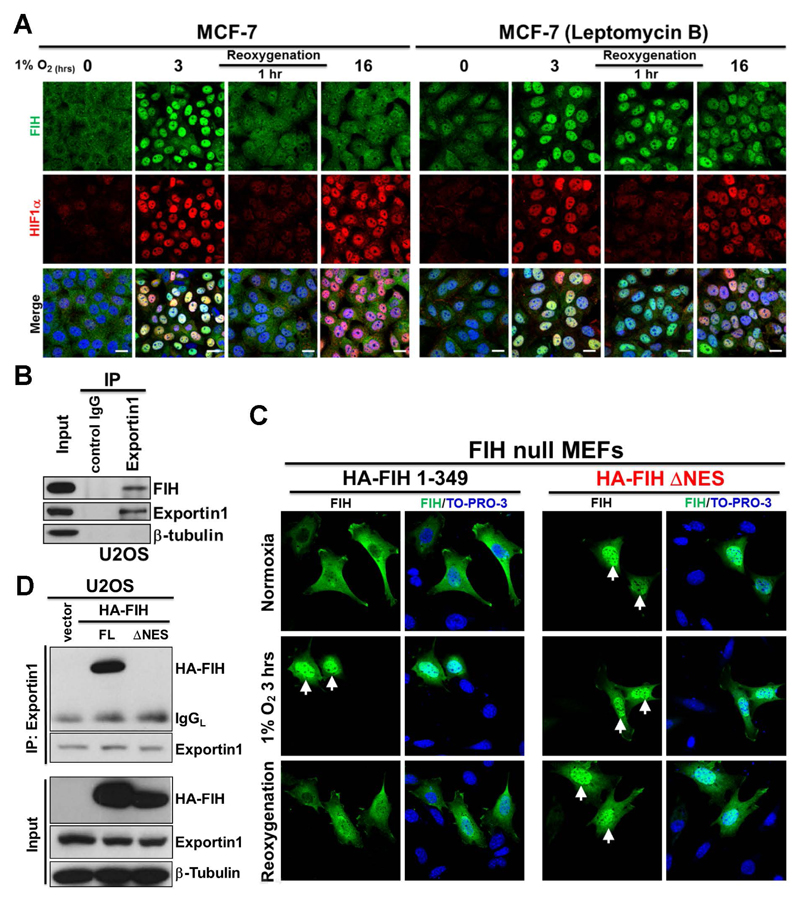
We observed that FIH was retained in the nucleus only for a few hours during application of hypoxia (Fig. 1B; Supplementary Fig. 1C). Interestingly, during re-oxygenation following hypoxia, FIH was observed in the nucleus for up to 1 hour whereas HIF1α was degraded more quickly (Supplementary Fig. 4A), suggesting that export of nuclear FIH is mediated by an independent process. Nuclear export of proteins is usually mediated by leucine-rich nuclear export signals (NES) that are recognized by nuclear export receptors (Tran et al., 2014). Seven exportins have been described so far, with chromosome region maintenance 1 (CRM1), also referred to as exportin1, being the most abundant (Tran et al., 2014). Leptomycin B is a potent and specific nuclear export inhibitor that inhibits exportin1 (Nishi et al., 1994). We found Leptomycin B treatment promoted the retention of FIH in the nucleus following 16 hours hypoxia and 1 hour of reoxygenation, while in non-treated cells, FIH was no longer detectable in the nucleus under these conditions (Fig. 4A). This indicates that FIH is exported by a Leptomycin B-sensitive pathway. Consistent with this, endogenous FIH and exportin1 co-immunoprecipitated in U2OS cells (Fig. 4B). Furthermore, a potential NES is predicted within FIH (residues 128-137) using NetNES (http://www.cbs.dtu.dk/services/NetNES/) (Supplementary Fig. 4B). We thus constructed a plasmid encoding hemagglutinin (HA)-tagged FIH lacking the predicted NES (HA-FIH ΔNES), and transfected wild type or ΔNES HA-tagged FIH plasmids into FIH null mouse embryonic fibroblasts (MEFs). Consistent with work described above we found that exogenously expressed wild type FIH was cytoplasmic, and that nuclear accumulation was observed upon hypoxia. Furthermore, after 1 hour re-oxygenation following hypoxia, FIH was cytoplasmic (Fig. 4C, HA-FIH 1-349 panel), consistent with our observation on endogenous FIH (Fig. 4A; Supplementary Fig. 4A). By contrast, exogenously expressed FIH lacking the predicted NES showed nuclear localization under all conditions (Fig. 4C, HA-FIH ΔNES panel). In addition, exogenously expressed FIH lacking the predicted NES failed to associate with exportin1 (Fig. 4D), highlighting the important role of this sequence in mediating nuclear export.
Figure 4. FIH exits nucleus via a Leptomycin B-sensitive exportin1 (CRM1)-dependent pathway.
(A) Immunofluorescence staining of FIH (green) and HIF1α (red) in MCF7 cells with the indicated hypoxia (0.5% O2) and reoxygenation treatments. TO-PRO-3 (blue) was used to stain nuclei. Scale bar: 20 µm.
(B) Total cell lysates from U2OS cells were immunoprecipitated with an anti-exportin1 antibody or control IgG. FIH, exportin1 and β-tubulin levels are indicated.
(C) Immunofluorescence staining of FIH (green) in FIH null mouse embryonic fibroblasts (MEFs) transfected with HA-FIH 1-349 or HA-FIH ΔNES followed by treatment in normoxia, hypoxia (1% O2) 3 hours or 1 hour reoxygenation. TO-PRO-3 (blue) was used to stain nuclei. Arrows: nuclear localization of signal.
(D) Total cell lysates from U2OS cells transfected with control vector, HA-FIH 1-349 (FL: full length) or HA-FIH ΔNES were immunoprecipitated with an anti-exportin 1 antibody. HA-FIH, exportin1 and β-tubulin levels are indicated. IgGL: IgG light chain.
These data demonstrate that FIH is exported via a Leptomycin B-sensitive exportin1 (CRM1)-dependent pathway; while upon hypoxia, FIH is actively imported by importin β1-mediated nuclear import in association with HIF1α.
Discussion
FIH was initially isolated as a negatively regulating factor of HIFα following a yeast two-hybrid screen using the final 251 residues of human HIF1α (576-826) as a bait (Mahon et al., 2001). The mechanism by which FIH represses HIFα transcriptional activity was not determined in this initial study. Independently, Lando et al. subsequently found that that an asparagine residue (N851 in mouse HIF2α, corresponding to N803 in human HIF1α), which is conserved in orthologous vertebrate HIF1α and HIF2α proteins, undergoes hydroxylation (Lando et al., 2002b). They demonstrated that asparaginyl hydroxylation is a mechanism for normoxic repression of the transcriptional activation by the HIF1α CAD via blocking the interaction of HIF with the CBP/p300 transcriptional coactivator proteins.
FIH was subsequently shown to be the HIF1α asparaginyl hydroxylase following bioinformatic analyses that predicted FIH to have a tertiary structure including a modified ‘double stranded β helix’ (DSBH) fold that is typical of the 2-OG-dependent hydroxylases (Hewitson et al., 2002; Lando et al., 2002a). This prediction was subsequently verified by crystallographic analyses (Elkins et al., 2003). Recombinant FIH was shown to catalyse the Fe (II) and 2-OG dependent C-3 hydroxylation of an asparagine residue in the CAD of HIFα isoforms (Hewitson et al., 2002; Lando et al., 2002a; McNeill et al., 2002).
FIH-catalysed HIFα hydroxylation blocks the interaction between the transcriptional coactivators/histone acetyl transferases p300/CBP and HIFα (Lando et al., 2002a). In vitro, p300 does not bind to HIFα CAD treated with wild type FIH, but does bind to HIFα CAD treated with a catalytically inactive FIH variant (Hewitson et al., 2002; Lando et al., 2002a). Subsequent work has also revealed that FIH accepts multiple substrates from the ankyrin repeat domain (ARD) family of proteins (Cockman et al., 2006; Cockman et al., 2009; Coleman et al., 2007; Janke et al., 2013; Karttunen et al., 2015; Zheng et al., 2008), which are located in different cellular compartments. As a result, intracellular processes that affect the subcellular location of FIH will determine its access to different substrates.
Our findings reveal dynamic mechanisms controlling the cellular localization of FIH. We employed both genetic and small-molecule interventions to identify a mechanism by which hypoxia induces nuclear translocation of FIH. Notably, we show that FIH accumulates in the nucleus within a short time window upon hypoxia treatment. We provide evidence that the presence of FIH in the nucleus is governed by two opposing processes; nuclear entry by “coupling” of FIH import with HIF1α for importin β1-mediated nuclear import and active export by a Leptomycin B-sensitive exportin1 (CRM1)-mediated nuclear export pathway. We identified a potential NES within FIH (residues 128-137) using bioinformatics and confirmed it by the comparison of NES-deleted FIH and full-length FIH under identical transfection in FIH null MEFs. We also report that nuclear import of FIH requires inhibition of its own enzyme activity. This is consistent with an earlier report by Cockman et al. showing that the FIH-substrate interactions could be stabilized by pre-treatment of cells with the catalytic inhibitor DMOG, most likely due to prolongation of otherwise transient interactions between enzyme and substrate (Cockman et al., 2009).
It is also notable that in addition to the HIF1α isoforms, FIH binds and hydroxylates a diverse array of ARD-containing proteins, including Notch (Coleman et al., 2007; Zheng et al., 2008), apoptosis-stimulating of p53 protein 2 (ASPP2) (Janke et al., 2013) and others (Cockman et al., 2006; Cockman et al., 2009; Karttunen et al., 2015). Unlike the very substantial effect of asparaginyl hydroxylation on HIF-mediated transcription, FIH-catalysed ARD-hydroxylation has not yet been found to have a clear role on ARD signalling (though it can affect ARD stability) (Kelly et al., 2009). These ARD-containing proteins, however, have a high affinity for FIH and are abundant in cells (Coleman et al., 2007; Wilkins et al., 2009), thus they compete with HIF1α for asparaginyl hydroxylation, which, as we have demonstrated in this study, is critical to facilitate the nuclear import of FIH. In addition, a group of ARD-containing protein use the RanGDP/AR (RaDAR) complex-mediated system for nuclear import (Lu et al., 2014). Interestingly, the Notch intracellular domain (NICD) has been reported to play a role in the nuclear accumulation of FIH in normoxic cells though the mechanism not explored (Zheng et al., 2008). Our findings may enable new insights into the paradox that FIH effectively hydroxylates HIF1α despite the presence of numerous and abundant competing ARD substrates. Competition between ARDs and HIFα for binding to FIH may not only directly regulate FIH-catalysed HIFα hydroxylation (as previously proposed) (Cockman et al., 2006; Schmierer et al., 2010), but also regulate the cytoplasmic versus nuclear localizations of FIH. The new data suggest a more complex and dynamic interface with the HIF transcriptional response, where FIH may be particularly important in modulating transitions between normoxic and hypoxic states. It is also important to note that FIH is a dimer (Elkins et al., 2003), thus has the potential to bind more than one protein (substrate) simultaneously, so potentially enabling further fine-tuning of the role of FIH in the hypoxic response.
The results shown in this study rationalize why several earlier studies failed to detect nuclear FIH under hypoxia (Linke et al., 2004; Metzen et al., 2003; Soilleux et al., 2005; Stolze et al., 2004), since the narrow window of the nuclear impact of FIH may have been missed. The results also potentially explain why, despite FIH being largely a cytoplasmic protein in normoxic cells, it has sometimes been observed in the nucleus in pathological tissues (Kroeze et al., 2010; Tan et al., 2007). A detailed understanding of the biological importance of cytoplasmic and nuclear FIH function may help to explain whether nuclear FIH causes or simply associates with pathological conditions.
Materials and methods
Cell culture, reagents and transfections
U2OS, HKe3, MCF7 cells and FIH null mouse embryonic fibroblasts (MEFs) were cultured in DMEM supplemented with 10% fetal bovine serum (Invitrogen) and antibiotics. All cells were kept at 37 °C and 10% CO2. No mycoplasma contamination was detected in the cell lines used. Hypoxic incubations were performed in In vivo2 400 hypoxic workstations (Ruskin Technologies). Chemicals DMOG, IOX1, IOX2, FG2216, VGB10B (IOX4), DFO and DM-NOFD were obtained or synthesized as reported (Chan et al., 2015; Chan et al., 2016; Chowdhury et al., 2013; Hopkinson et al., 2013; McDonough et al., 2005; Mole et al., 2003; Yeh et al., 2017).
siRNA oligos against the genes encoding for FIH (MU-004073-02-0002), HIF1α (MU-004018-05-0002), HIF2α (MU-004814-01-0002), HIF1β (MU-007207-01-0002) and Importin β1 (MU-017523-01-0002) were purchased from Dharmacon. Sequences are available from Dharmacon, or on request. As a negative control we used siGENOME RISC-Free siRNA (Dharmacon). Cells were transfected with the indicated siRNA oligos at a final concentration of 35 nM using DharmaFECT 1 reagent (Dharmacon).
Wild-type HA-tagged FIH (HA-FIH 1-349) plasmid was cloned as described previously (Coleman et al., 2007). HA-tagged FIH plasmid lacking the predicted NES 128-137 (HA-FIH ΔNES) was cloned by the site-directed mutagenesis. Transfections were performed with FuGENE 6 (Promega), according to the manufacturer's instructions.
Western blot analysis
Western blot analysis was carried out with lysates from cells with urea buffer (8 M urea, 1 M thiourea, 0.5% CHAPS, 50 mM dithiothreitol and 24 mM spermine). For preparation of cytoplasmic and nuclear proteins, NE-PER nuclear and cytoplasmic extraction reagents (Pierce) were used in accordance with the manufacturer’s protocol. Cytoplasmic and nuclear fractions were isolated from U2OS cells with indicated treatment, and β-tubulin was used as a loading control for the cytoplasmic fraction, whereas PARP was used as a loading control for the nuclear fraction. For immunoprecipitations, the cells were lysed for 30 min at 4 °C in pNAS buffer (50 mM Tris-HCl at pH 7.5, 120 mM NaCl, 1 mM EDTA and 0.1% Nonidet P-40), with protease inhibitors. Cell extracts were then precleared with protein G beads and incubated with antibodies against Importin β1 (2 µl per mg of protein lysate, Cell Signaling Technology, 8673, rabbit polyclonal), exportin1 (2 µg per mg of protein lysate, Sigma-Aldrich, E7784, rabbit polyclonal), HIF1α (2 µg per mg of protein lysate, Novus Biologicals, NB100-479, rabbit polyclonal) or p300 (2 µg per mg of protein lysate, Millipore, 05-257, mouse monoclonal RW128) for 16 h at 4 °C. Immunoprecipitates were washed four times with cold PBS followed by the addition of SDS sample buffer. The bound proteins were separated on SDS–polyacrylamide gels and subjected to immunoblotting with the indicated antibodies.
Primary antibodies were from Novus Biologicals (HIF1α, 1:1,000, NB100-479, rabbit polyclonal), Abcam (β-tubulin, 1:5,000, ab6046, rabbit polyclonal; GAPDH, 1:2,000, ab9385, rabbit polyclonal; Ku80, 1:2,000, ab80592, rabbit monoclonal EPR3468), Sigma-Aldrich (exportin1, 1:1,000, E7784, rabbit polyclonal), BD Transduction Laboratories (HIF1α, 1:1,000, 610958, mouse monoclonal Clone 54/HIF1α), Cell Signaling Technology (PARP, 1:1,000, 9542, rabbit polyclonal; HIF1β, 1:1,000, 5537, rabbit monoclonal D28F3; importin β1, 1:1,000, 8673, rabbit polyclonal), Santa Cruz Biotechnology (HA, 1:1,000, sc-7392, mouse monoclonal Clone F-7), HIF2α (1:50, clone-190b) (Wiesener et al., 1998), HIF1α hydroxy-Asn803 (N803OH) (1:5,000, mouse monoclonal) (Lee et al., 2008) and FIH (1:200, mouse monoclonal 162C) (Stolze et al., 2004). Signals were detected using an ECL detection system (GE Healthcare) and evaluated by ImageJ 1.42q software (National Institutes of Health).
Immunofluorescence microscopy
Cells were fixed in 4% PBS–paraformaldehyde for 15 min, incubated in 0.1% Triton X-100 for 5 min on ice, then in 0.2% fish skin gelatin in PBS for 1 h and stained for 1 h with an anti-FIH antibody (1:50, mouse monoclonal 162C)(Stolze et al., 2004), anti-HIF1α N803OH (1:500, mouse monoclonal) (Lee et al., 2008) and anti-HIF1α antibody (1:100, BD Transduction Laboratories 610958, mouse monoclonal Clone 54/HIF1α or 1:100, Novus Biologicals NB100-479, rabbit polyclonal). Protein expression was detected using Alexa Fluor (1:400, Molecular Probes) for 20 min. DAPI or TO-PRO-3 (Invitrogen) was used to stain nucleic acids (1:1,000). Samples were observed using a confocal microscope system (LSM 510 or LSM 710; Carl Zeiss). Acquired images were analysed by ImageJ 1.42q software (National Institutes of Health) using in-house plugins written for quantification of nuclear signal. Briefly, four high power fields were selected for analysis of each treatment. To avoid being biased by the FIH staining, each field was selected by viewing nuclear (DAPI) staining only to identify near-confluent cells and thereby maximise the cell numbers (~ 100 cells) included in the analysis. For each high power field, binary image masks were created of FIH and DAPI positive staining to define regions of interest (ROI) for analysis. The DAPI staining mask was used to define the nuclear ROI, which was then applied, by the image calculator, to the original FIH staining images to isolate nuclear staining within each image. Using the image calculator, the DAPI mask was subtracted from the FIH mask to create a staining mask defining the non-nuclear ROI. Quantitative fluorescence data were exported from ImageJ generated histograms into Microsoft Excel software for further analysis and presentation. Cells with the ratio of nuclear FIH mean intensity over non-nuclear FIH mean intensity bigger than 2 are considered as nuclear FIH positive.
Statistical analysis and repeatability of experiments
Each experiment was repeated at least twice. Unless otherwise noted, data are presented as mean ± s.d., and a two-tailed, unpaired Student’s t-test was used to compare two groups for independent samples. P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
This work was supported by the Ludwig Institute for Cancer Research Ltd, the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC), and the Academy of Medical Sciences/the Wellcome Trust Springboard Award [SBF002\1038] (YW). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. Prof Schofield thanks the Wellcome Trust and Cancer Research UK for support. We are grateful to Dr. Senji Shirasawa for providing HKe3 cells and to Dr. Myung Kyu Lee for the HIF1α hydroxy-Asn803(N803OH) antibody. We thank Mark Shipman, Hokfung Chan and Richard Lisle for their technical assistance in image analysis.
Footnotes
Competing Financial Interests
PJR and CJS are scientific co-founders and hold equity in ReOx Ltd,. A University spin-out company aiming to develop HIF hydroxylase inhibitors.
References
- Adams RL, Wente SR. Uncovering nuclear pore complexity with innovation. Cell. 2013;152:1218–21. doi: 10.1016/j.cell.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Rout MP. The yeast nuclear pore complex and transport through it. Genetics. 2012;190:855–83. doi: 10.1534/genetics.111.127803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 2003;22:4082–90. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken CP, Fedele AO, Linke S, Balrak W, Lisy K, Whitelaw ML, Peet DJ. Cell-specific regulation of hypoxia-inducible factor(HIF)-1alpha and HIF-2alpha stabilization and transactivation in a graded oxygen environment. J Biol Chem. 2006;281:22575–85. doi: 10.1074/jbc.M600288200. [DOI] [PubMed] [Google Scholar]
- Chan MC, Atasoylu O, Hodson E, Tumber A, Leung IK, Chowdhury R, Gomez-Perez V, Demetriades M, Rydzik AM, Holt-Martyn J, et al. Potent and Selective Triazole-Based Inhibitors of the Hypoxia-Inducible Factor Prolyl-Hydroxylases with Activity in the Murine Brain. PLoS One. 2015;10:e0132004. doi: 10.1371/journal.pone.0132004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MC, Ilott NE, Schodel J, Sims D, Tumber A, Lippl K, Mole DR, Pugh CW, Ratcliffe PJ, Ponting CP, et al. Tuning the Transcriptional Response to Hypoxia by Inhibiting Hypoxia-inducible Factor (HIF) Prolyl and Asparaginyl Hydroxylases. J Biol Chem. 2016;291:20661–73. doi: 10.1074/jbc.M116.749291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilov D, Camenisch G, Kvietikova I, Ziegler U, Gassmann M, Wenger RH. Induction and nuclear translocation of hypoxia-inducible factor-1 (HIF-1): heterodimerization with ARNT is not necessary for nuclear accumulation of HIF-1alpha. J Cell Sci. 1999;112(Pt 8):1203–12. doi: 10.1242/jcs.112.8.1203. [DOI] [PubMed] [Google Scholar]
- Chowdhury R, Candela-Lena JI, Chan MC, Greenald DJ, Yeoh KK, Tian YM, McDonough MA, Tumber A, Rose NR, Conejo-Garcia A, et al. Selective small molecule probes for the hypoxia inducible factor (HIF) prolyl hydroxylases. ACS Chem Biol. 2013;8:1488–96. doi: 10.1021/cb400088q. [DOI] [PubMed] [Google Scholar]
- Cockman ME, Lancaster DE, Stolze IP, Hewitson KS, McDonough MA, Coleman ML, Coles CH, Yu X, Hay RT, Ley SC, et al. Posttranslational hydroxylation of ankyrin repeats in IkappaB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor inhibiting HIF (FIH) Proc Natl Acad Sci U S A. 2006;103:14767–72. doi: 10.1073/pnas.0606877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockman ME, Webb JD, Kramer HB, Kessler BM, Ratcliffe PJ. Proteomics-based identification of novel factor inhibiting hypoxia-inducible factor (FIH) substrates indicates widespread asparaginyl hydroxylation of ankyrin repeat domain-containing proteins. Mol Cell Proteomics. 2009;8:535–46. doi: 10.1074/mcp.M800340-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ML, McDonough MA, Hewitson KS, Coles C, Mecinovic J, Edelmann M, Cook KM, Cockman ME, Lancaster DE, Kessler BM, et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J Biol Chem. 2007;282:24027–38. doi: 10.1074/jbc.M704102200. [DOI] [PubMed] [Google Scholar]
- Dann CE, 3rd, Bruick RK, Deisenhofer J. Structure of factor-inhibiting hypoxia-inducible factor 1 An asparaginyl hydroxylase involved in the hypoxic response pathway. Proc Natl Acad Sci U S A. 2002;99:15351–6. doi: 10.1073/pnas.202614999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depping R, Jelkmann W, Kosyna FK. Nuclear-cytoplasmatic shuttling of proteins in control of cellular oxygen sensing. J Mol Med (Berl) 2015;93:599–608. doi: 10.1007/s00109-015-1276-0. [DOI] [PubMed] [Google Scholar]
- Depping R, Steinhoff A, Schindler SG, Friedrich B, Fagerlund R, Metzen E, Hartmann E, Kohler M. Nuclear translocation of hypoxia-inducible factors (HIFs): involvement of the classical importin alpha/beta pathway. Biochim Biophys Acta. 2008;1783:394–404. doi: 10.1016/j.bbamcr.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Elkins JM, Hewitson KS, McNeill LA, Seibel JF, Schlemminger I, Pugh CW, Ratcliffe PJ, Schofield CJ. Structure of factor-inhibiting hypoxia-inducible factor (HIF) reveals mechanism of oxidative modification of HIF-1 alpha. J Biol Chem. 2003;278:1802–6. doi: 10.1074/jbc.C200644200. [DOI] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Hewitson KS, McNeill LA, Riordan MV, Tian YM, Bullock AN, Welford RW, Elkins JM, Oldham NJ, Bhattacharya S, Gleadle JM, et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J Biol Chem. 2002;277:26351–5. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- Hopkinson RJ, Tumber A, Yapp C, Chowdhury R, Aik W, Che KH, Li XS, Kristensen JBL, King ONF, Chan MC, et al. 5-Carboxy-8-hydroxyquinoline is a Broad Spectrum 2-Oxoglutarate Oxygenase Inhibitor which Causes Iron Translocation. Chem Sci. 2013;4:3110–3117. doi: 10.1039/C3SC51122G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke K, Brockmeier U, Kuhlmann K, Eisenacher M, Nolde J, Meyer HE, Mairbaurl H, Metzen E. Factor inhibiting HIF-1 (FIH-1) modulates protein interactions of apoptosis-stimulating p53 binding protein 2 (ASPP2) J Cell Sci. 2013;126:2629–40. doi: 10.1242/jcs.117564. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Kang J, Shin SH, Yoon H, Huh J, Shin HW, Chun YS, Park JW. FIH is an oxygen sensor in ovarian cancer for G9a/GLP-driven epigenetic regulation of metastasis-related genes. Cancer Res. 2017 doi: 10.1158/0008-5472.CAN-17-2506. [DOI] [PubMed] [Google Scholar]
- Karttunen S, Duffield M, Scrimgeour NR, Squires L, Lim WL, Dallas ML, Scragg JL, Chicher J, Dave KA, Whitelaw ML, et al. Oxygen-dependent hydroxylation by FIH regulates the TRPV3 ion channel. J Cell Sci. 2015;128:225–31. doi: 10.1242/jcs.158451. [DOI] [PubMed] [Google Scholar]
- Kelly L, McDonough MA, Coleman ML, Ratcliffe PJ, Schofield CJ. Asparagine beta-hydroxylation stabilizes the ankyrin repeat domain fold. Mol Biosyst. 2009;5:52–8. doi: 10.1039/b815271c. [DOI] [PubMed] [Google Scholar]
- Kiriakidis S, Henze AT, Kruszynska-Ziaja I, Skobridis K, Theodorou V, Paleolog EM, Mazzone M. Factor-inhibiting HIF-1 (FIH-1) is required for human vascular endothelial cell survival. FASEB J. 2015;29:2814–27. doi: 10.1096/fj.14-252379. [DOI] [PubMed] [Google Scholar]
- Kroeze SG, Vermaat JS, van Brussel A, van Melick HH, Voest EE, Jonges TG, van Diest PJ, Hinrichs J, Bosch JL, Jans JJ. Expression of nuclear FIH independently predicts overall survival of clear cell renal cell carcinoma patients. Eur J Cancer. 2010;46:3375–82. doi: 10.1016/j.ejca.2010.07.018. [DOI] [PubMed] [Google Scholar]
- Kuzmanov A, Wielockx B, Rezaei M, Kettelhake A, Breier G. Overexpression of factor inhibiting HIF-1 enhances vessel maturation and tumor growth via platelet-derived growth factor-C. Int J Cancer. 2012;131:E603–13. doi: 10.1002/ijc.27360. [DOI] [PubMed] [Google Scholar]
- Lancaster DE, McNeill LA, McDonough MA, Aplin RT, Hewitson KS, Pugh CW, Ratcliffe PJ, Schofield CJ. Disruption of dimerization and substrate phosphorylation inhibit factor inhibiting hypoxia-inducible factor (FIH) activity. Biochem J. 2004;383:429–37. doi: 10.1042/BJ20040735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002a;16:1466–71. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002b;295:858–61. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- Lee SH, Jeong Hee M, Eun Ah C, Ryu SE, Myung Kyu L. Monoclonal antibody-based screening assay for factor inhibiting hypoxia-inducible factor inhibitors. J Biomol Screen. 2008;13:494–503. doi: 10.1177/1087057108318800. [DOI] [PubMed] [Google Scholar]
- Liang K, Ding XQ, Lin C, Kang YJ. Featured Article: Hypoxia-inducible factor-1alpha dependent nuclear entry of factor inhibiting HIF-1. Exp Biol Med (Maywood) 2015;240:1446–51. doi: 10.1177/1535370215570821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke S, Stojkoski C, Kewley RJ, Booker GW, Whitelaw ML, Peet DJ. Substrate requirements of the oxygen-sensing asparaginyl hydroxylase factor-inhibiting hypoxia-inducible factor. J Biol Chem. 2004;279:14391–7. doi: 10.1074/jbc.M313614200. [DOI] [PubMed] [Google Scholar]
- Lu M, Zak J, Chen S, Sanchez-Pulido L, Severson DT, Endicott J, Ponting CP, Schofield CJ, Lu X. A code for RanGDP binding in ankyrin repeats defines a nuclear import pathway. Cell. 2014;157:1130–45. doi: 10.1016/j.cell.2014.05.006. [DOI] [PubMed] [Google Scholar]
- Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–86. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marxsen JH, Stengel P, Doege K, Heikkinen P, Jokilehto T, Wagner T, Jelkmann W, Jaakkola P, Metzen E. Hypoxia-inducible factor-1 (HIF-1) promotes its degradation by induction of HIF-alpha-prolyl-4-hydroxylases. Biochem J. 2004;381:761–7. doi: 10.1042/BJ20040620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough MA, McNeill LA, Tilliet M, Papamicael CA, Chen QY, Banerji B, Hewitson KS, Schofield CJ. Selective inhibition of factor inhibiting hypoxia-inducible factor. J Am Chem Soc. 2005;127:7680–1. doi: 10.1021/ja050841b. [DOI] [PubMed] [Google Scholar]
- McNeill LA, Hewitson KS, Claridge TD, Seibel JF, Horsfall LE, Schofield CJ. Hypoxia-inducible factor asparaginyl hydroxylase (FIH-1) catalyses hydroxylation at the beta-carbon of asparagine-803. Biochem J. 2002;367:571–5. doi: 10.1042/BJ20021162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzen E, Berchner-Pfannschmidt U, Stengel P, Marxsen JH, Stolze I, Klinger M, Huang WQ, Wotzlaw C, Hellwig-Burgel T, Jelkmann W, et al. Intracellular localisation of human HIF-1 alpha hydroxylases: implications for oxygen sensing. J Cell Sci. 2003;116:1319–26. doi: 10.1242/jcs.00318. [DOI] [PubMed] [Google Scholar]
- Mole DR, Schlemminger I, McNeill LA, Hewitson KS, Pugh CW, Ratcliffe PJ, Schofield CJ. 2-oxoglutarate analogue inhibitors of HIF prolyl hydroxylase. Bioorg Med Chem Lett. 2003;13:2677–80. doi: 10.1016/s0960-894x(03)00539-0. [DOI] [PubMed] [Google Scholar]
- Myllyharju J. Prolyl 4-hydroxylases, master regulators of the hypoxia response. Acta Physiol (Oxf) 2013;208:148–65. doi: 10.1111/apha.12096. [DOI] [PubMed] [Google Scholar]
- Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem. 1994;269:6320–4. [PubMed] [Google Scholar]
- Pelletier J, Dayan F, Durivault J, Ilc K, Pecou E, Pouyssegur J, Mazure NM. The asparaginyl hydroxylase factor-inhibiting HIF is essential for tumor growth through suppression of the p53-p21 axis. Oncogene. 2012;31:2989–3001. doi: 10.1038/onc.2011.471. [DOI] [PubMed] [Google Scholar]
- Peng H, Hamanaka RB, Katsnelson J, Hao LL, Yang W, Chandel NS, Lavker RM. MicroRNA-31 targets FIH-1 to positively regulate corneal epithelial glycogen metabolism. FASEB J. 2012a;26:3140–7. doi: 10.1096/fj.11-198515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Kaplan N, Hamanaka RB, Katsnelson J, Blatt H, Yang W, Hao L, Bryar PJ, Johnson RS, Getsios S, et al. microRNA-31/factor-inhibiting hypoxia-inducible factor 1 nexus regulates keratinocyte differentiation. Proc Natl Acad Sci U S A. 2012b;109:14030–4. doi: 10.1073/pnas.1111292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe PJ. HIF-1 and HIF-2: working alone or together in hypoxia? J Clin Invest. 2007;117:862–5. doi: 10.1172/JCI31750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe PJ. Oxygen sensing and hypoxia signalling pathways in animals: the implications of physiology for cancer. J Physiol. 2013;591:2027–42. doi: 10.1113/jphysiol.2013.251470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J, Pilkington R, Garcia Munoz A, Nguyen LK, Rauch N, Kennedy S, Monsefi N, Herrero A, Taylor CT, von Kriegsheim A. Substrate-Trapped Interactors of PHD3 and FIH Cluster in Distinct Signaling Pathways. Cell Rep. 2016;14:2745–60. doi: 10.1016/j.celrep.2016.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmierer B, Novak B, Schofield CJ. Hypoxia-dependent sequestration of an oxygen sensor by a widespread structural motif can shape the hypoxic response--a predictive kinetic model. BMC Syst Biol. 2010;4:139. doi: 10.1186/1752-0509-4-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz CC, Rodriguez J, Pickel C, Burr S, Fabrizio JA, Nolan KA, Spielmann P, Cavadas MA, Crifo B, Halligan DN, et al. FIH Regulates Cellular Metabolism through Hydroxylation of the Deubiquitinase OTUB1. PLoS Biol. 2016;14:e1002347. doi: 10.1371/journal.pbio.1002347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim J, Cowburn AS, Palazon A, Madhu B, Tyrakis PA, Macias D, Bargiela DM, Pietsch S, Gralla M, Evans CE, et al. The Factor Inhibiting HIF Asparaginyl Hydroxylase Regulates Oxidative Metabolism and Accelerates Metabolic Adaptation to Hypoxia. Cell Metab. 2018;27:898–913 e7. doi: 10.1016/j.cmet.2018.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soilleux EJ, Turley H, Tian YM, Pugh CW, Gatter KC, Harris AL. Use of novel monoclonal antibodies to determine the expression and distribution of the hypoxia regulatory factors PHD-1, PHD-2, PHD-3 and FIH in normal and neoplastic human tissues. Histopathology. 2005;47:602–10. doi: 10.1111/j.1365-2559.2005.02280.x. [DOI] [PubMed] [Google Scholar]
- Stolze IP, Tian YM, Appelhoff RJ, Turley H, Wykoff CC, Gleadle JM, Ratcliffe PJ. Genetic analysis of the role of the asparaginyl hydroxylase factor inhibiting hypoxia-inducible factor (FIH) in regulating hypoxia-inducible factor (HIF) transcriptional target genes [corrected] J Biol Chem. 2004;279:42719–25. doi: 10.1074/jbc.M406713200. [DOI] [PubMed] [Google Scholar]
- Tan EY, Campo L, Han C, Turley H, Pezzella F, Gatter KC, Harris AL, Fox SB. Cytoplasmic location of factor-inhibiting hypoxia-inducible factor is associated with an enhanced hypoxic response and a shorter survival in invasive breast cancer. Breast Cancer Res. 2007;9:R89. doi: 10.1186/bcr1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian YM, Yeoh KK, Lee MK, Eriksson T, Kessler BM, Kramer HB, Edelmann MJ, Willam C, Pugh CW, Schofield CJ, et al. Differential sensitivity of hypoxia inducible factor hydroxylation sites to hypoxia and hydroxylase inhibitors. J Biol Chem. 2011;286:13041–51. doi: 10.1074/jbc.M110.211110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, King MC, Corbett AH. Macromolecular transport between the nucleus and the cytoplasm: Advances in mechanism and emerging links to disease. Biochim Biophys Acta. 2014;1843:2784–2795. doi: 10.1016/j.bbamcr.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Bu F, Royer C, Serres S, Larkin JR, Soto MS, Sibson NR, Salter V, Fritzsche F, Turnquist C, et al. ASPP2 controls epithelial plasticity and inhibits metastasis through beta-catenin-dependent regulation of ZEB1. Nat Cell Biol. 2014;16:1092–104. doi: 10.1038/ncb3050. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ngo VN, Marani M, Yang Y, Wright G, Staudt LM, Downward J. Critical role for transcriptional repressor Snail2 in transformation by oncogenic RAS in colorectal carcinoma cells. Oncogene. 2010;29:4658–70. doi: 10.1038/onc.2010.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesener MS, Turley H, Allen WE, Willam C, Eckardt KU, Talks L, Wood SM, Gatter KC, Harris AL, Pugh CW, et al. Induction of endothelial PAS domain protein-1 by hypoxia: characterization and comparison with hypoxia-inducible factor-1alpha. Blood. 1998;92:2260–8. [PubMed] [Google Scholar]
- Wilkins SE, Hyvarinen J, Chicher J, Gorman JJ, Peet DJ, Bilton RL, Koivunen P. Differences in hydroxylation and binding of Notch and HIF-1alpha demonstrate substrate selectivity for factor inhibiting HIF-1 (FIH-1) Int J Biochem Cell Biol. 2009;41:1563–71. doi: 10.1016/j.biocel.2009.01.005. [DOI] [PubMed] [Google Scholar]
- Yeh TL, Leissing TM, Abboud MI, Thinnes CC, Atasoylu O, Holt-Martyn JP, Zhang D, Tumber A, Lippl K, Lohans CT, et al. Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials. Chem Sci. 2017;8:7651–7668. doi: 10.1039/c7sc02103h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Fu Z, Linke S, Chicher J, Gorman JJ, Visk D, Haddad GG, Poellinger L, Peet DJ, Powell F, et al. The asparaginyl hydroxylase factor inhibiting HIF-1alpha is an essential regulator of metabolism. Cell Metab. 2010;11:364–78. doi: 10.1016/j.cmet.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Linke S, Dias JM, Gradin K, Wallis TP, Hamilton BR, Gustafsson M, Ruas JL, Wilkins S, Bilton RL, et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc Natl Acad Sci U S A. 2008;105:3368–73. doi: 10.1073/pnas.0711591105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.