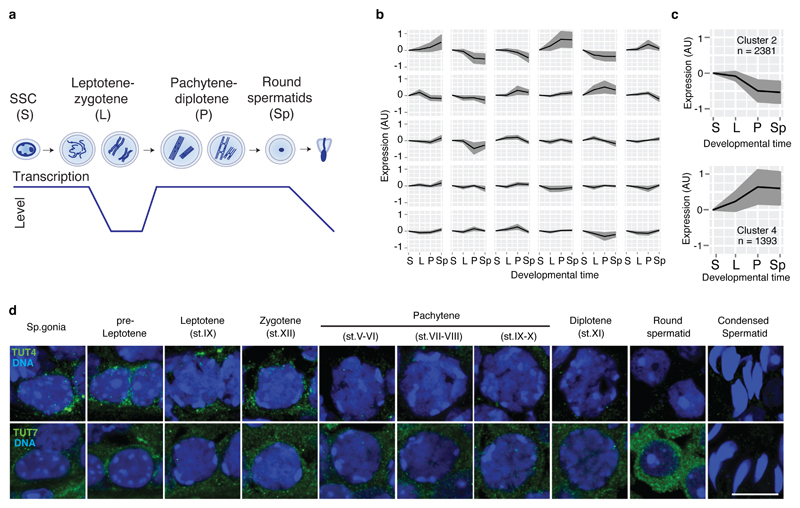
Figure 1. A programmed wave of RNA degradation takes place during the leptotene-zygotene to pachytene transition.
a, Schematic representation of different spermatogenic transitions. Spermatogonial stem cells (SSC; S), leptotene-zygotene (L), pachytene-diplotene (P) and round spermatids (Sp). Levels of transcriptional activity are indicated below. b, Expression change across different spermatogenic transitions as defined in a for clusters of transcripts with similar expression profiles. Clusters were generated using the Markov clustering algorithm. The black line indicates the mean expression of the group, and the gray area indicates the standard deviation. c, Expression changes as defined in b for the two clusters showing changes in gene expression during the leptotene-zygotene to pachytene transition (n, number of genes). d, Confocal immunofluorescence micrographs of different spermatogenic cell types are shown. Testes from WT animals were stained with an antibody against TUT4 (green), upper panel. Testes from Tut7HA-GFP/HA-GFP mice were stained with an anti-HA antibody (green), lower panel. DNA was stained with Hoechst 33342 (blue). Scale bar, 10 μm.