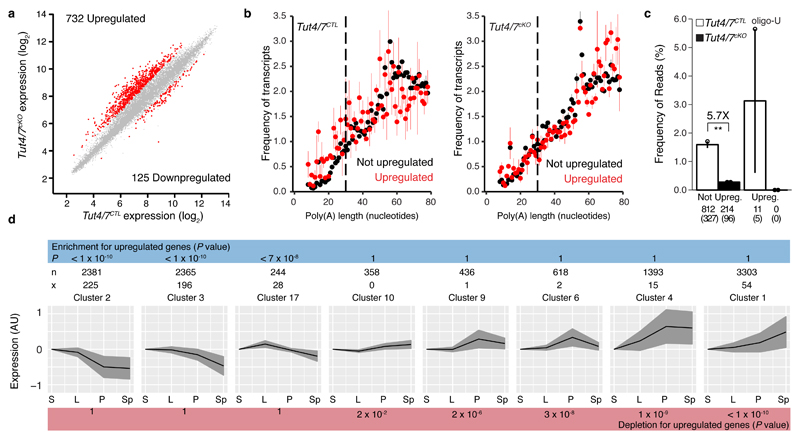
Figure 5. TUT4/7 are required for clearance of transcripts in pachytene spermatocytes.
a, Scatter plot of pachytene mRNA expression levels in Tut4/7CTL vs Tut4/7cKO animals. Transcripts significantly changed (P < 0.01 and fold-change > 2, Moderated t-statistic adjusted; Tut4/7CTL, n=4; Tut4/7cKO, n=3) are highlighted in red. The numbers of significantly upregulated and downregulated genes are indicated. b, Poly(A) tail length distribution for pachytene mRNAs from Tut4/7CTL and Tut4/7cKO mice. The distribution is shown for upregulated (red) or not upregulated (black) transcripts. Each dot represents the mean value of two biological replicates and, the error bars indicate the range. The dotted vertical line at 30 nucleotides separates short and long tails. c, Mono- and oligo-uridylation of pachytene mRNAs with short poly(A) tails from Tut4/7CTL and Tut4/7cKO mice. Each dot represents a biological replicate. The height of the bar and the error bars represent the mean and range, respectively. The fold change between groups is indicated together with its significance (**P < 0.01, t-test two-sided; Tut4/7CTL, n=2; Tut4/7cKO, n=2). The number of transcripts and genes (in brackets) is shown for each group. d, Enrichment analysis of upregulated transcripts in Tut4/7cKO pachytene cells across different clusters of genes group according to their expression across spermatogenesis. The expression profile of each cluster is shown. The black line indicates the mean expression of the group, and the gray area indicates the standard deviation. The number of transcripts in a cluster (n) and the number of upregulated transcripts in that cluster (x) are shown for each cluster. The P values (Hypergeometric test) for enrichment or depletion are also indicated. (S, spermatogonia stem cells; L, leptotene-zygotene; P, pachytene-diplotene, and Sp, round spermatids).