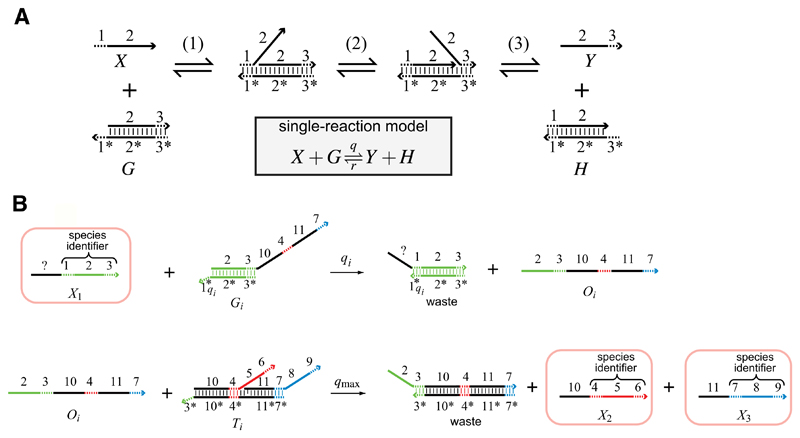
Fig. 4.
A 4-domain scheme for implementing chemical reaction networks in DNA, reproduced from Soloveichik et al. (2010). a An elementary DNA strand displacement interaction, modelled by the chemical reaction X + G → Y + H. Chemical species X denotes a single DNA strand consisting of domains 1 and 2, where each domain corresponds to a DNA sequence. The strand X is written <1 2>, where the 3’ end of the strand is assumed to be on the right, represented graphically by an arrowhead. The species G denotes a complex consisting of strand <2 3> hybridized to the strand <3* 2* 1*>, where the star (*) denotes Watson-Crick complementarity. The reaction takes place in three steps: (1) The domain 1 binds to its complement 1*. The reaction is reversible, since the domain 1 is assumed to be short enough to spontaneously unbind. These short domains are referred to as toeholds; (2) The domain 2 of strand <1 2> displaces the domain 2 of strand <2 3> by a random walk process, referred to as branch migration; (3) The toehold domain 3 spontaneously unbinds from its complement 3*. b A 4-domain encoding of the formal unimolecular reaction X1 → X2 + X3, with reaction index i. The species of this formal reaction are represented as DNA strands and highlighted by boxes, with X1 represented as < ? 1 2 3>, X2 as <10 4 5 6> and X3 as <11 7 8 9>. Black domains are assumed to be unique to each reaction i, while green, red and blue domains are associated to species X1, X2 and X3, respectively. The formal reaction X1 → X2 + X3 is implemented by two DNA complexes, which are assumed to be present in excess and are consumed over time. The first complex G1 binds the species X1 and produces the intermediate Oi, while the second complex Ti binds the intermediate Oi and produces the two species X2 and X3. The additional intermediate step is needed to ensure that the reactant species X1 does not contain any overlapping domains with the product species X2 and X3. Note that the sequence of toehold domain 1qi can also be adjusted to be only partially complementary to domain 1, in order to tune the reaction rate qi. (Color figure online)