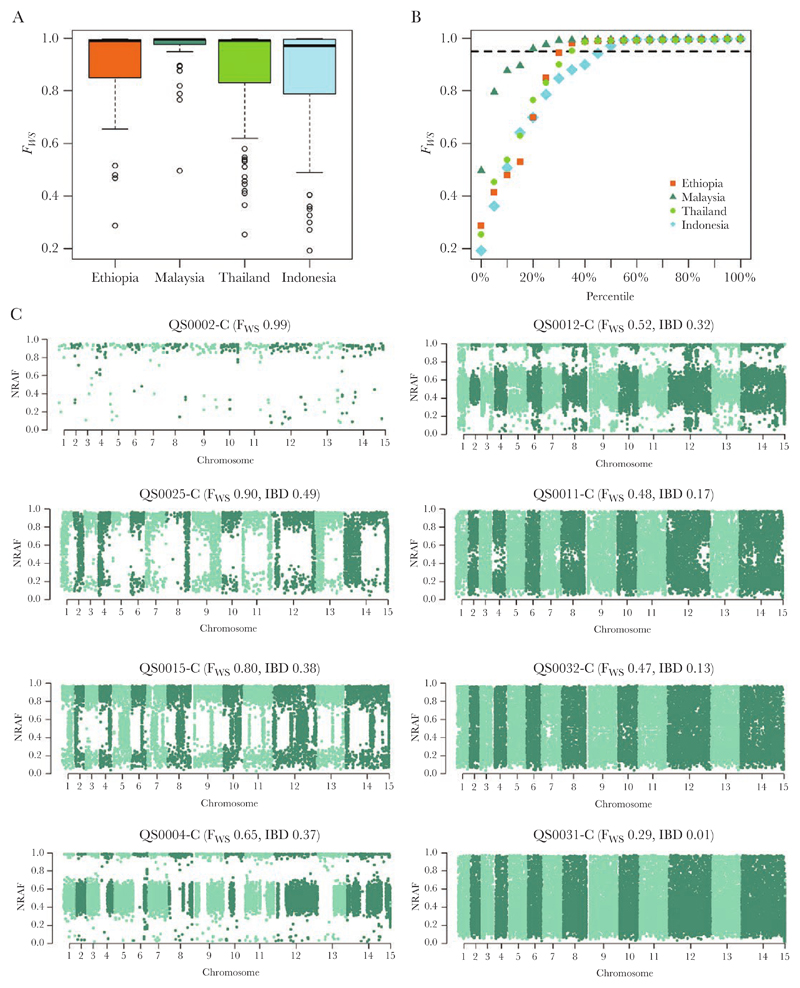
Figure 1.
Within-sample infection complexity in Ethiopia relative to the Southeast Asian populations and within polyclonal Ethiopian infections. The boxplots (A) and scatterplots (B) illustrate the distribution of within-sample F statistic (FWS) scores in Ethiopia relative to Thailand, Indonesia, and Malaysia. Data are presented on all 293 high-quality samples. B, Dashed line illustrates FWS = 0.95, above which infections are essentially monoclonal. Ethiopia exhibits an intermediate proportion of monoclonal infections relative to Malaysia and Indonesia, most comparable to Thailand. C, Manhattan plots of the nonreference allele frequency (NRAF) in 7 Ethiopian infections identified as polyclonal based on FWS < 0.95, and 1 monoclonal infection as a baseline reference (QS0002-C). Trends in the pairwise identity by descent (IBD) between clones in a given infection (as determined by DEploid-IBD) correlated positively with the FWS scores and approximated runs of homozygosity (RoH, not presented). DEploid-IBD found evidence of 2 major clones in QS0025-C, QS0015-C, and QS0004-C, and 3 clones in QS0012-C, QS0011-C, QS0032-C, and QS0031-C. The clones within QS0025-C demonstrated the highest pairwise IBD (0.49), indicative of siblings sharing approximately 50% of their genomes, as illustrated by the long stretches of homology (regions with NRAF approximating 0 or 1) on each chromosome. In striking contrast, the clones within QS0031-C exhibited the lowest pairwise IBD (<0.01%), with no evidence of recent common ancestry, rather reflecting a probable superinfection.