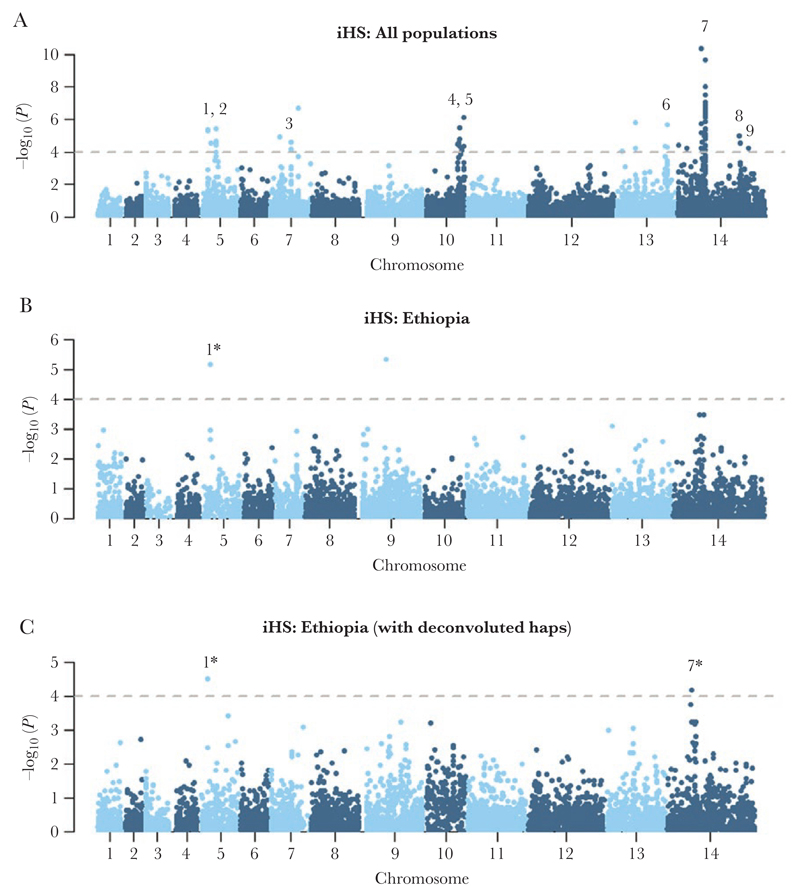
Figure 3.
Genome-wide scans of extended haplotype homozygosity using the integrated haplotype score (iHS) illustrating regions under recent directional selection in Ethiopia and across populations. Manhattan plots of the iHS P value for the given populations: all monoclonal (within-sample F statistic [FWS] > 0.95) samples from Ethiopia, Thailand, and Indonesia (A), all monoclonal (FWS > 0.95) samples from Ethiopia (B), and all monoclonal (FWS > 0.95) samples from Ethiopia plus the 7 major haplotypes derived from polyclonal Ethiopian isolates that were deconvoluted (C) using DEploid identity by descent (IBD) software. Data are presented on 260 982 loci for which derived alleles could be confidently called. The dashed black lines demark the thresholds of –log10 (P value) > 4: signals supported by a minimum of 3 single-nucleotide polymorphisms (SNPs) above the threshold within 50 kb of one another and with an overall SNP density <10 kb per SNP are numbered. Details can be found in Supplementary Data 4. In brief, the putative genetic drivers include an SPRY domain protein (PVP01_0508000) (signal 1), 3 conserved proteins with unknown function (PVP01_0515000, PVP01_1029200, and PVP01_1455300) (signals 2, 4, and 9 respectively), gamma-glutamylcysteine synthetase (PVP01_0717300), an AP2 domain transcription factor (PVP01_1418100) or ferredoxin (PVP01_1419000) (signal 7), and an amino acid transporter (PVP01_1449600) (signal 8). The driver in the 92-kb region on chromosome 13 (signal 6) remains unclear. *Signals were supported by a minimum of 3 SNPs above the threshold in the population-wide data but only 1 SNP in Ethiopia.