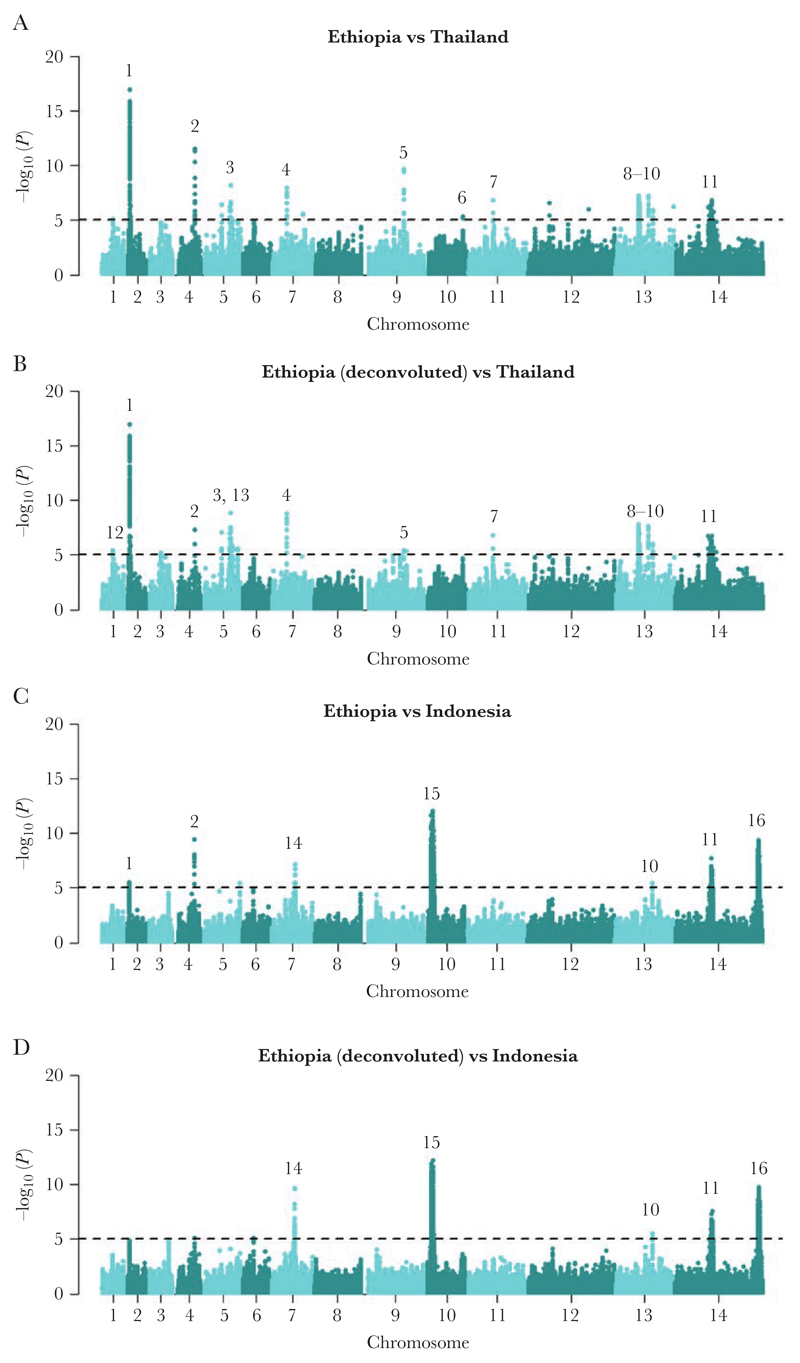
Figure 4.
Genome-wide scans of Rsb-based cross-population extended haplotype homozygosity illustrating regions under divergent selection between Ethiopia and Thailand and Indonesia. A–D, Manhattan plots of the Rsb P value for the given populations. The dashed black lines demark the thresholds of –log10 (P value) > 5: signals supported by a minimum of 3 single-nucleotide polymorphisms (SNPs) above the threshold within 50 kb of one another and with an overall SNP density <10 kb per SNP are numbered. The multi-SNP signals are detailed in Supplementary Data 4. Several previously described signals in known or putative drug resistance candidates were identified including MRP1 (signal 1), DHFR (signal 3), and DHPS (signal 11). Signals were also observed in regions upstream of CRT (signal 12) and MDR1 (signal 15). The putative genetic drivers in other regions include MSP5 (signal 2), a tRNA (signal 4), AMA1 (signal 5), a merozoite surface protein 3 family cluster (signal 6), a voltage-dependent anion-selective channel (signal 8), liver-specific protein 1 (signal 10), and an AP2 domain transcription factor (signal 13). The putative genetic drivers in regions 14 and 16 are conserved Plasmodium proteins with unknown function, and those in regions 7 and 9 remain unclear.