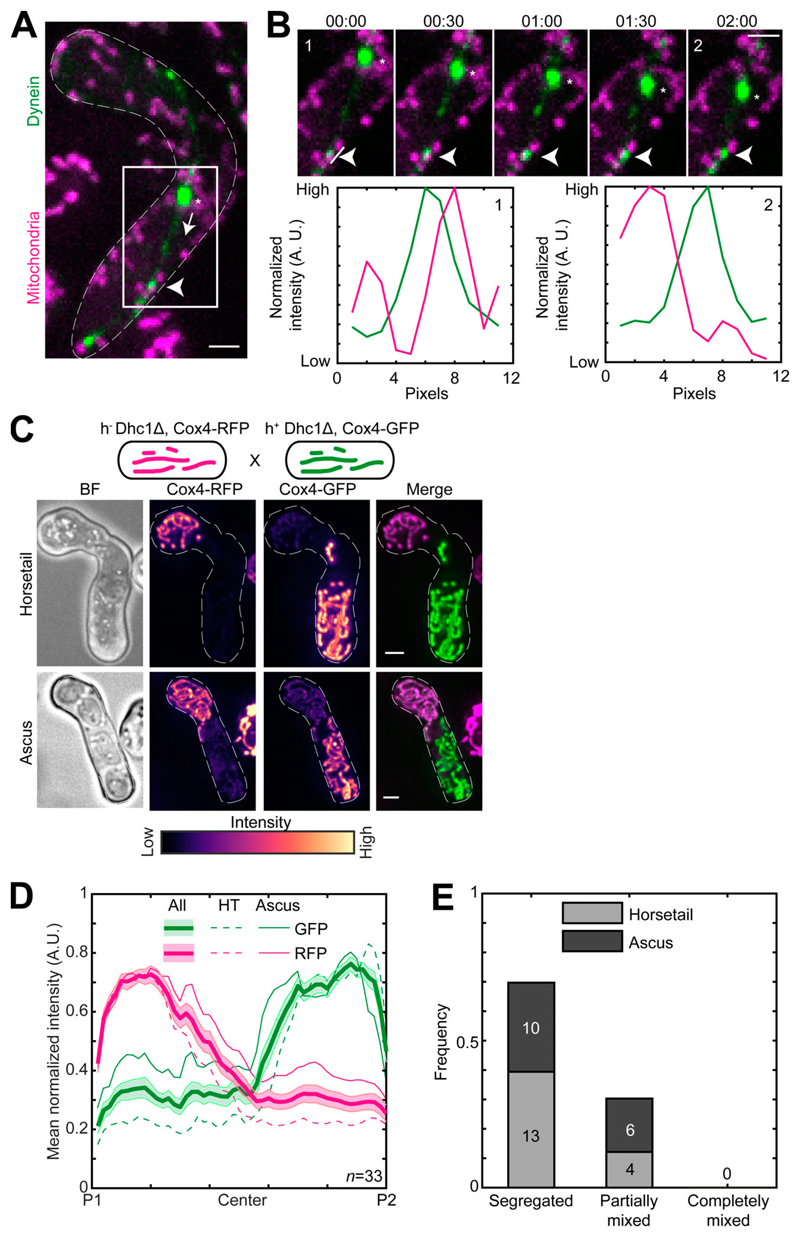
Figure 5. Dynein and mitochondria do not bind to the same Mcp5 foci.
(A) Maximum-intensity–projected images showing a merge of dynein (green) and mitochondria (magenta) in a meiotic cell undergoing nuclear oscillations (left, strain VA099; see Table S1). The white arrowhead points to a representative dynein spot on the cortex, the asterisk indicates the position of the SPB, and the dashed arrow points to the direction of SPB movement. (B) Montage of the inset indicated in A with dynein in green and mitochondria in magenta (top) and plots of normalized intensity (bottom) of dynein (green) and mitochondria (magenta) 1 pixel to the left of the line indicated in montage numbered 1 (left) and numbered 2 (right). The white arrowheads point to the dynein spot. Time is indicated above the images of the montage in minutes:seconds. (C) Schematic of the cross performed (top, strain VA091xVA092; see Table S1), maximum-intensity–projected images of the bright-field channel (BF; first from left), mitochondria labeled with Cox4-RFP (second from left) and mitochondria labeled with Cox4-GFP (third from left) represented in the intensity map to the bottom of the images, and their merge (right) during the early stage (horsetail, top) and late stage (ascus, bottom) of meiosis. (D) Plot of mean normalized intensities of RFP (magenta lines) and GFP (green lines) in the horsetail stage (HT; dashed lines, n = 17), ascus stage (Ascus; solid lines, n= 16), and the stages combined (All; thick solid lines) across the length of the cell from the cross indicated in C (n = 33). Shaded regions represent SEM. (E) Stacked bar plot of frequency of segregated, partially mixed and completely mixed phenotypes observed in horsetail (light gray) and ascus (dark gray) stages from the data in D. In A–C, scale bars represent 2 µm and dashed lines represent cell outlines.