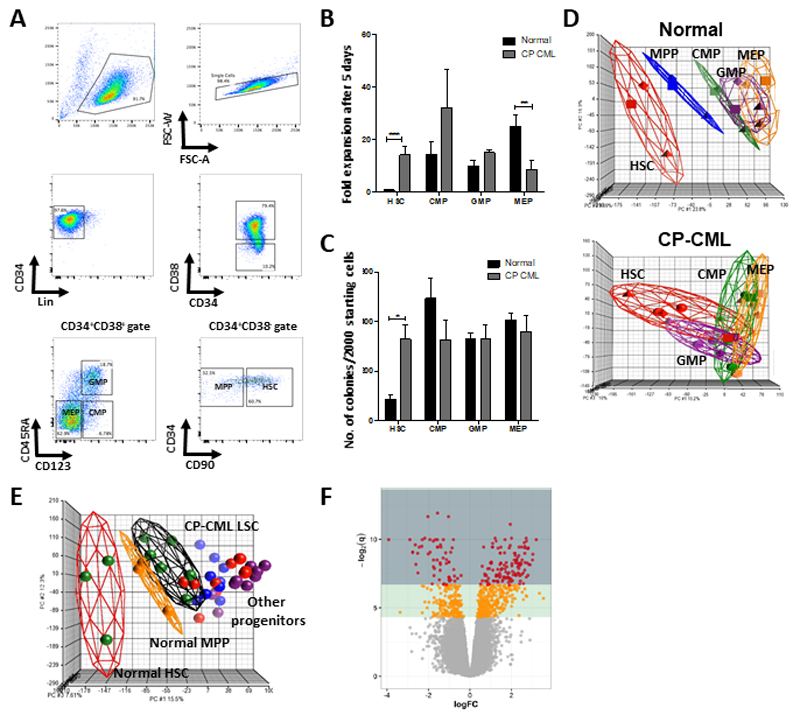
Figure 1. CML LSCs are more proliferative than normal HSCs.
(A) A representative sorting strategy of CP-CML samples is depicted. (B) Fold expansion after 5 days (mean +/- standard deviation) are shown (**, p<0.01; ***, p<0.001). (C) Colony counts are represented (mean +/- standard deviation) per 2000 starting cells to allow comparison between groups (*, p<0.05). (D) Principal component analysis (PCA) of the gene expression profiles from GSE47927 was used to identify global differences between normal and CP-CML. An individual point within the axes represents each microarray and each subpopulation grouping by a different colour. Separate PCAs of normal and CP-CML subpopulations. (E) PCA of normal and CP samples overlapped. (F) A volcano plot from the same dataset showing deregulation of genes in a comparison of Lin-CD34+CD38-CD90+ cells from 6 CP-CML samples compared to 3 normal samples (GSE47927). Areas of significance are indicated by a dark green (equivalent to q<=0.01) or light green (equivalent to q<=0.05) background. Genes reaching significance are coloured red (q<=0.01) or orange (q<=0.05); non-significant genes are grey.