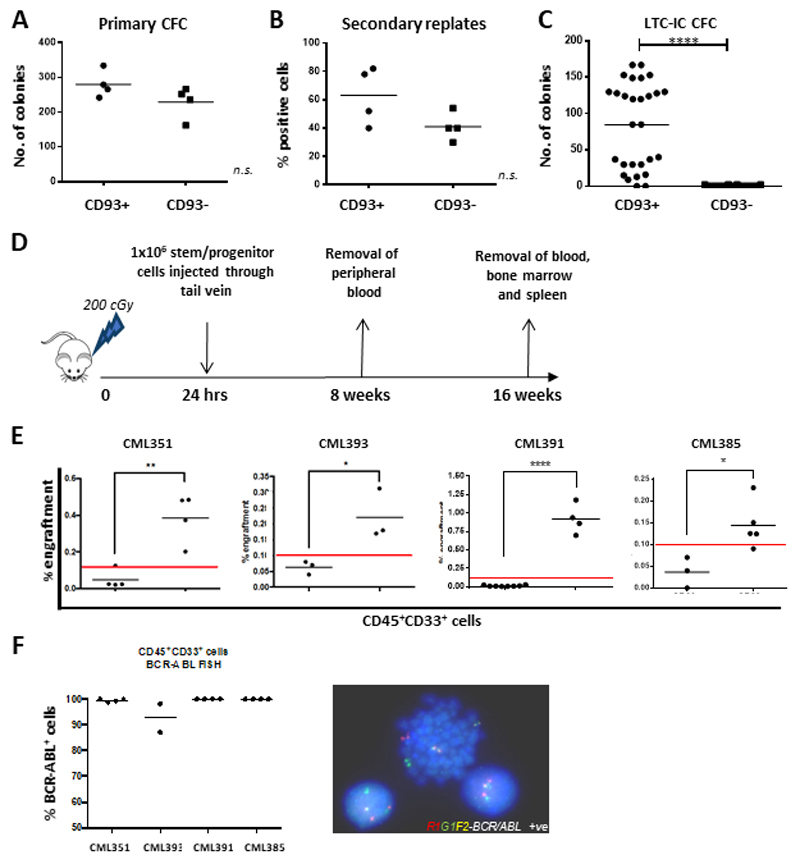
Figure 3. CD93+, but not CD93-, CP-CML cells have stem cell characteristics in vitro and in vivo.
(A) CP-CML samples (n=4) were sorted according to figure S2 and plated in duplicate into Methocult for 10-12 days prior to colony counts. The graph indicates the mean number of colonies for each CP-CML sample, with mean and standard deviation for each population shown. (B) 50 primary colonies per sample were replated in Methocult in 96 well plates and incubated at 37 degrees, 5% CO2 for 12 days before positive wells were counted. The graph indicates the percentage of positive wells per population (n=4 CP-CML samples), with mean and standard deviation shown. (C) The graph indicates the number of colonies from each experimental arm (n=3 CP-CML samples), with mean and standard deviation shown. (D) Representation of in vivo experimental model. CP-CML Lin-CD34+CD93+ or Lin-CD34+CD93- cells were isolated by FACS sorting (1x106 cells/mouse), washed and transplanted via tail vein injection into sub-lethally irradiated (2 Gy) 8-12 week-old NSG mice. PB was sampled at 8 weeks to assess for CD45+33+ expression. All mice were euthanized after 16 weeks and marrow contents of femurs were obtained. Cells were labelled with anti-human CD45, CD33 and CD19 antibodies prior to analysis by flow cytometry. This allowed for analysis of lineages within the engrafted samples. (E) Human myeloid cell engraftment was characterized by percentage of CD45+CD33+ cells. (F) Human CD45+ cells were isolated by FACS sorting and analyzed by FISH for the BCR-ABL gene rearrangement. Percentage of BCR-ABL positive cells, as determined from analysis of a minimum of 100 cells, was assessed for each murine experiment.