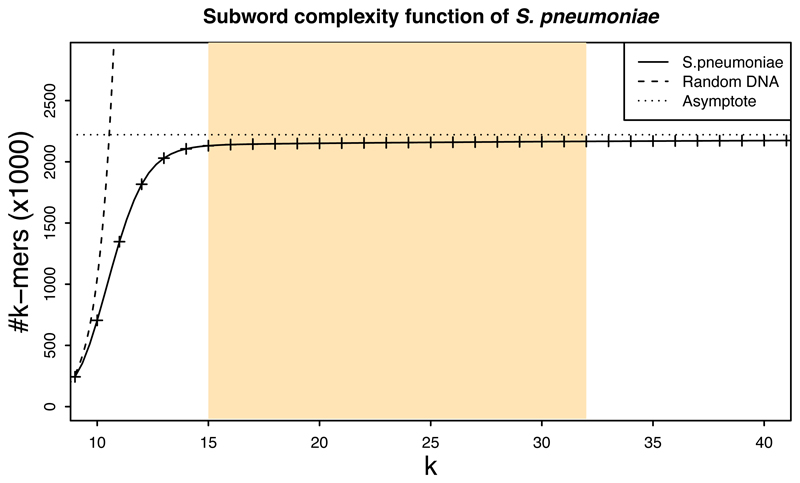
Extended Data Fig. 5. Subword complexity of pneumococcus.
The plot depicts the number of canonical k-mers as a function of k for S. pneumoniae ATCC 700669 (GenBank accession: ‘NC_011900.1’) and for a random DNA text containing all possible k-mers. For k<10, the pneumococcus k-mer composition is similar to the one of random text. For k>14, the k-mer sets are almost saturated and the complexity grows very slowly. Since the genome length is finite and bacterial chromosomes are circular, the function attains its maximum at the genome size (2,221,315 in this case). The highlighted region corresponds to the range of values of k, which are suitable for use in RASE.