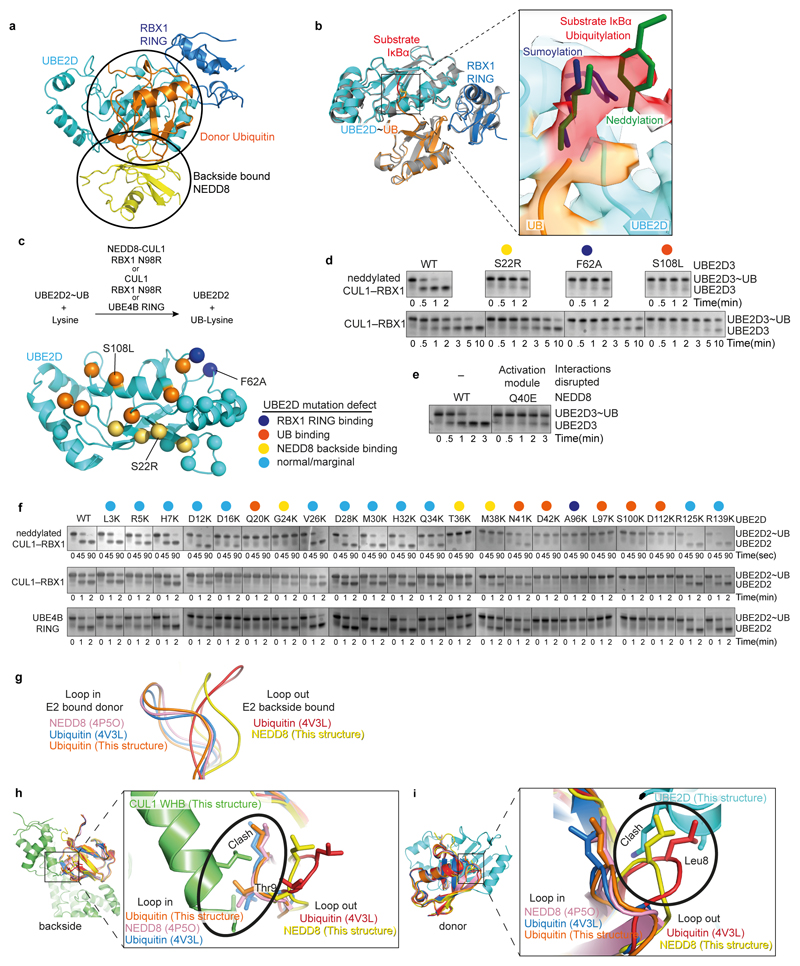
Extended Data Figure 7. Interactions shaping the catalytic architecture of neddylated CRL1β-TRCP-UBE2D~UB-IκBα substrate intermediate.
a, NEDD8 and the catalytic module from the structure representing the neddylated CRL1β-TRCP-UBE2D~UB-IκBα intermediate, highlighting distinctive interactions between NEDD8 (yellow) and donor UB (orange) with UBE2D. b, Catalytic module from the neddylated CRL1β-TRCP-UBE2D~UB-IκBα intermediate, highlighting the covalently-linked proxy for the IκBα substrate’s acceptor in the active site relative to a superimposed representative prior crystal structure of an isolated RING-UBE2D~UB complex (grey, PDB: 4AP4)28,29. In the inset, the density for the covalently-linked proxy for IκBα substrate’s acceptor is shown in red the active site. The chemical trap superimposes with consensus acceptors visualized in active sites of sumoylation63 and neddylation39 intermediates, where aromatic side-chains guide the lysine targets (blue and green, respectively)39,73. However, UBE2D’s myriad substrates neither conform to a specific motif, nor do they or UBE2D display specific side-chains guiding lysine acceptors into the catalytic center. Instead, in the neddylated CRL1β-TRCP-UBE2D~UB-substrate complex, density from backbone atoms preceding the chemical proxy for the acceptor Lys corresponds to the aromatic guides in sumoylation and neddylation intermediates. c, Overview of assays for activation of intrinsic reactivity of UBE2D~UB intermediate. Top, schematic of pulse-chase assay for testing effects of UBE2D mutations on activation, monitoring UBE2D~UB discharge to free lysine activated by neddylated CUL-RBX1 compared to unneddylated or RING-like UBE4B controls. Bottom, sites of mutations shown as spheres on structure of UBE2D from cryo EM structure of neddylated CRL1β-TRCP-UBE2D~UB-substrate complex. Sphere colors reflect both the locations and the effects on UBE2D~UB discharge to free lysine. Sites of mutations with marginal or no effect are shown in cyan, whereas those with major effects are colored. Mutations causing major defects map to RBX1 RING-binding site (blue), the interaction surface with the donor UB (orange), and the interaction surface with NEDD8 (yellow). d, Representative Coommassie-stained SDS-PAGE gels (of two independent experiments) shown for reactions monitoring substrate-independent discharge of UBE2D~UB to free lysine, in presence of CUL1-RBX1 (N98R) that was either neddylated or unneddylated (K720R),with either WT or indicated UBE2D3 mutants at binding sites for backside-bound NEDD8 (S22R), the RBX1 RING (F62A), and the covalently-linked donor UB in the closed conformation (S108L). e, Same as in d except testing effect of NEDD8 Q40E, which would disrupt the activation module. f, Reactions performed as in d, except with indicated variants of UBE2D2, in reactions with CUL1-RBX1 (N98R) that was either neddylated or unneddylated (K720R), or with the optimized RING-like U-box domain from UBE4B as a reference58. For mutations reporting on the catalytic conformation (G24K, T36K, M38K, A96K and D112K), representative gels are shown for two experiments. All other experiments were performed once. g, Comparison of β1/β2 -loop conformations after superimposing the indicated structures of NEDD8 and UB. The comparison suggests that while NEDD8 and UB can adopt both “loop in” and “loop out” conformations, donors linked to E2 active sites in RING activated complexes adopt the “loop in” conformation, and those bound to UBE2D backside adopt “loop out” conformations. h, Only a “loop out” conformation is compatible with the neddylated CRL activation module structure, because “loop in” conformation from the structures shown in g would prevent noncovalent interactions with CUL1 WHB domain (green). i, Only a “loop out” conformation is compatible for the CUL1-linked NEDD8 to bind the catalytic module, because “loop in” conformations in the structures shown in g would prevent noncovalent interactions with the UBE2D backside (cyan).