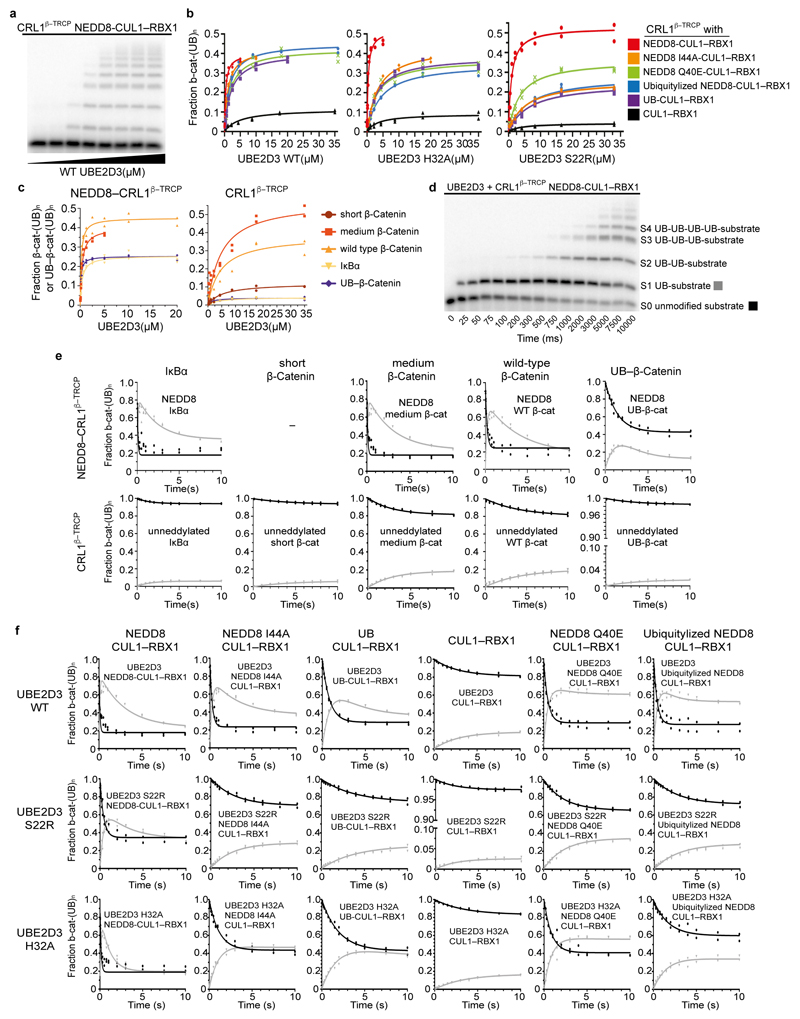
Extended Data Figure 1. Quantitative pre-steady state enzyme kinetics of neddylated CRL1β-TRCP- and UBE2D-dependent ubiquitylation.
Gel images are representative of independent technical replicates (n=2); The symbols from graphs show the data from independent experiments (n = 2), a, Autoradiogram of SDS-PAGE gel showing products of ubiquitylation reactions under single encounter conditions for interaction of radiolabeled substrate (“medium β-catenin” substrate peptide derived from β-catenin) with neddylated CRL1β-TRCP, titrating UBE2D3 (referred to as UBE2D hereafter in legend). Each lane represented a single ubiquitylation reaction that was used to estimate the fraction of peptide that had been converted into ubiquitylated products as a function of UBE2D concentration. b, Plots of the fraction of substrate that had been converted to ubiquitylated products versus UBE2D concentration for ubiquitylation reactions containing either WT (as shown in a) or UBE2D S22R or H32A mutants. Various CRL1β-TRCP complexes were assayed that contained either WT neddylated CRL1β-TRCP (red), CRL1β-TRCP complexes modified by NEDD8 variants harboring I44A (orange), Q40E (green), “Ubiquitylizing” (L2Q K4F E14T D16E G63K G64E) substitutions (blue), CRL1β-TRCP modified by UB R72A that is competent for ligation to CUL1 (purple), or unmodified CRL1β-TRCP (CUL1 with the neddylation site K720R mutation, black). Duplicate data points from independent experiments performed with identical samples are shown and were fit to the Michaelis-Menten model to estimate the K m of UBE2D for CRL1β-TRCP using non-linear curve fitting (GraphPad Prism). c, Plots of the fraction of substrate that had been converted to ubiquitylated products versus UBE2D concentration for ubiquitylation reactions with various substrate peptides: derived from IκBα (but with a single acceptor Lys); derived from β-catenin; derived from β-catenin with different spacing between the phosphodegron motif and a potential acceptor Lys (a “medium β-catenin” substrate peptide with a 9-residue spacer between the β-catenin phosphodegron and acceptor matching the relative position of these moieties in IκBα, and “short β-catenin” substrate wherein the four residues between these moieties are too few to bridge the structurally-observed gap between the substrate receptor and UBE2D~UB active site); and a homogeneous UB linked-β-catenin generated by sortase-mediated transpeptidation wherein the only lysines are from UB. d, Autoradiogram of SDS-PAGE gel showing results from rapid quench-flow reactions under pre-steady state single encounter conditions for interaction of radiolabeled substrate (a “medium β-catenin” phosphopeptide) with CRL1β-TRCP. The representative raw data are from a reaction using WT UBE2D and WT neddylated CRL1β-TRCP and show time-resolved conjugation of increasing numbers of individual UB molecules. S0 = substrate with 0 UBs, S1 = substrate with 1 UB, S2 = substrate with 2 UBs, etc. e, Plots comparing various substrate peptides described in c, showing disappearance of unmodified substrate (S0) with black circles, and the appearance of mono-ubiquitylated substrate (S1) with gray triangles, in rapid quench-flow reactions all performed as in d and under single encounter conditions as in a. Duplicate data points from independent experiments performed with identical samples are shown. The data were fit to closed form equations (Mathematica) as previously described59 to obtain both the rates for the transfer of the first UB to substrate (k obs S0-S1) and of the second UB to the singly UB-modified substrate (k obs S1-S2) as well as their associated standard error (Table 1). f, Plots from experiments performed and analyzed as described in e, except with radiolabeled “medium β-catenin” peptide substrate, CRL1β-TRCP variants containing the indicated versions of CUL1-RBX1, and with either WT or indicated mutant versions of UBE2D.