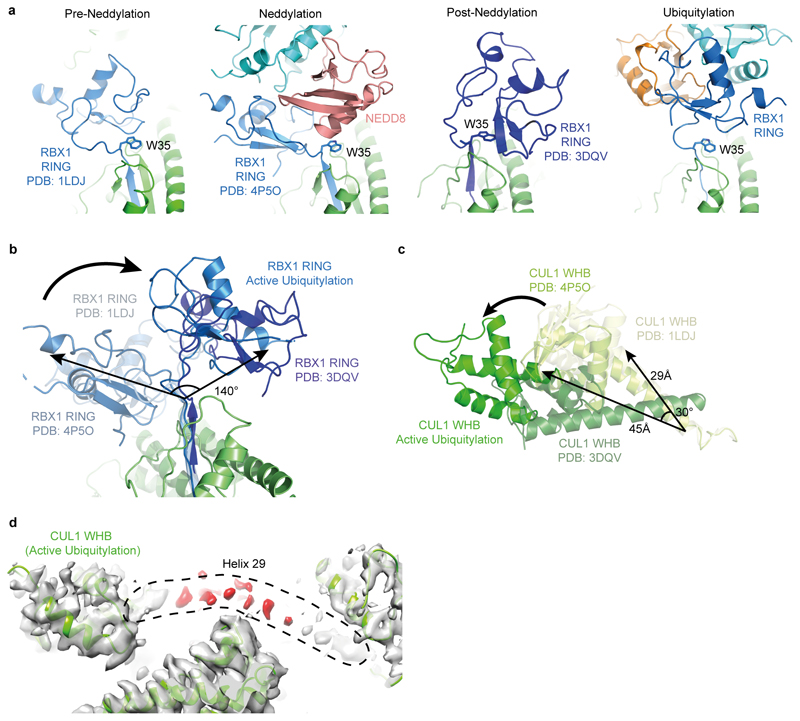
Extended Data Figure 5. Extraordinary cullin-RING conformational changes in catalytic architecture juxtaposing substrate and ubiquitylation active site.
a, Side-by-side comparison of relative RING domain locations in different CRL complexes after superposition of the C/R domains from the original CUL1-RBX1 structure (PDB ID 1LDJ, “Pre-Neddylation”, which data herein shows is dynamic, although the crystal structure likely captured the conformation allowing CAND1 binding and substrate receptor exchange)7, the structure representing the Neddylation reaction (PDB ID 4P5O)39, and a structure of a neddylated CUL5-RBX1 domain (PDB ID 3DQV, labeled “Post-Neddylation”, which revealed potential for neddylated CUL WHB and RBX1 RING domain conformational changes3, and data herein shows is dynamic), and the structure presented here showing how the neddylated CUL1 WHB domain and RBX1 RING are harnessed in a catalytic architecture for “Active ubiquitylation”. RBX1’s Trp35 is highlighted to show it serving as a multifunctional platform for either the RING domain in different orientations, or for the E2-linked NEDD8 during neddylation39. b, Superposition of the structures shown in a, highlighting different relative RING positions. c, Comparison of CUL WHB domain relative locations after superimposing their C/R domains (not shown). d, Cryo EM density from the neddylated CRL1β-TRCP-UBE2D~UB-substrate intermediate complex, showing patchiness of region corresponding to CUL1 “Helix-29”7. This CUL1 region connecting the C/R and WHB domains is visible only as patchy density, whereas in prior cullin crystals this forms the rod-like Helix-29 continuing into the WHB domain7. It seems CUL1’s Helix-29 dissolves into a flexible tether, which rationalizes the previously observed proteolytic sensitivity of this region in a neddylated CUL1-RBX1 complex3, and enables the displacement and rotation required for placing the ensuing WHB domain and its linked NEDD8 at the center of the ubiquitylation complex.