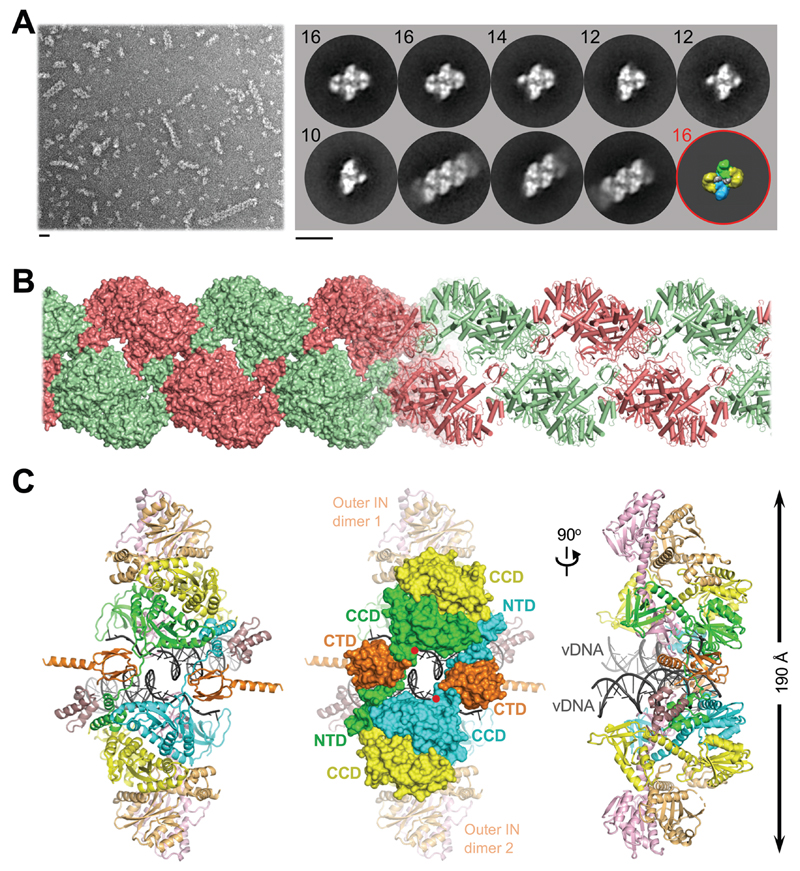
Fig. 1. Reconstruction of the SIVrcm intasome core.
(A) Raw image (left) and 2D class averages (right) of negatively stained SIVrcm intasome particles; apparent numbers of IN subunits are indicated for non-stacked classes. Particle distributions are given in fig. S4. The envelope of the hexadecameric maedi-visna virus intasome (red circle; central and flanking IN tetramers in blue/green and yellow, respectively) is shown for comparison; scale bars are 0.2 nm. (B) Atomistic reconstruction of the SIVrcm intasome stack shown as space fill (left) and cartoons (right); separate repeat units are shown in alternating red and green colors. (C) Detailed view of a single intasomal repeat representing a pair of viral DNA ends (vDNA, grey cartoons) synapsed between a pair of IN tetramers (composed of yellow, orange, pink, and either green or cyan IN protomers; the active sites of the green and cyan molecules (red dots) catalyze DNA recombination). The repeat unit is completed by pairs of C-terminal (orange) and N-terminal (dark magenta) domains donated by IN chains belonging to neighboring repeats. These CTDs are critical to form the conserved intasome core (CIC), which is shown in space fill mode in the middle panel. CCD, catalytic core domain.