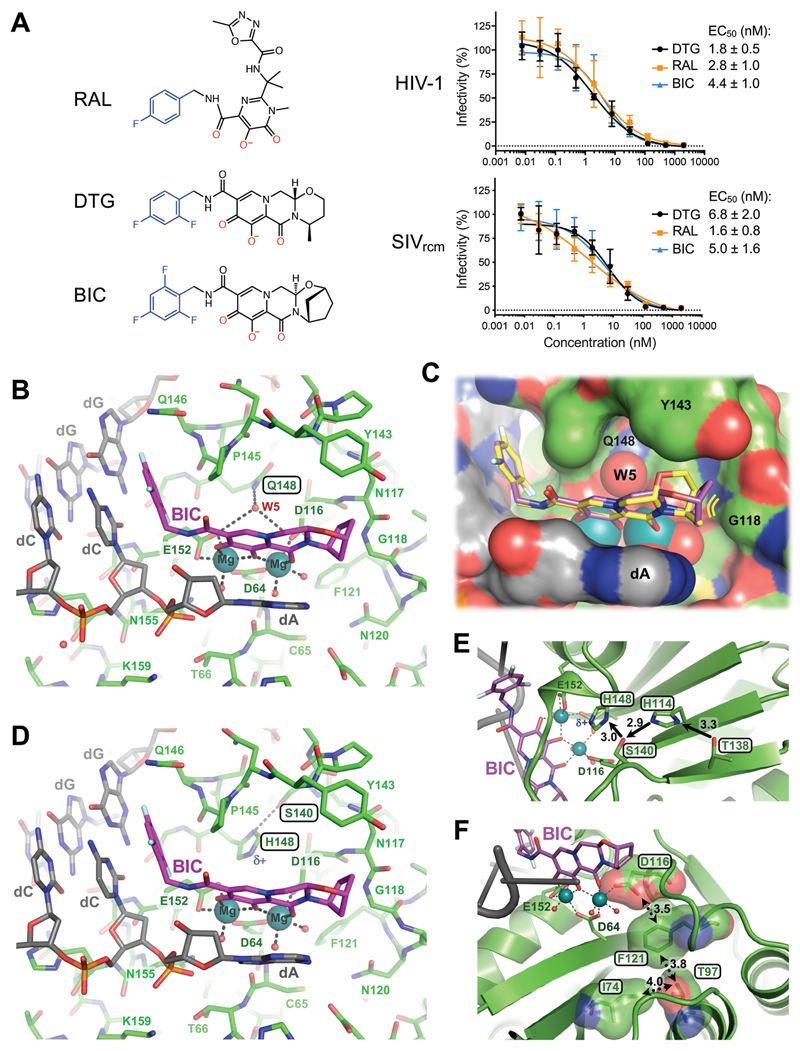
Fig. 2. Binding modes of second generation INSTIs in the IN active site.
(A) Chemical structures of select first- (RAL) and second-generation (DTG, BIC) INSTIs (left; halo-benzyl groups in blue and metal-chelating oxygen atoms in red) and viral sensitivities (right). Results are averages and standard deviations of minimally n = 2 experiments, with each experiment conducted in triplicate; EC50 values are noted. (B) Active site of the SIVrcm intasome in complex with BIC; protein, DNA, and drug are shown as sticks. Blue spheres are Mg2+ ions, water molecules are shown as small red spheres. (C) Superposition of BIC (magenta) and DTG (yellow) bound structures with protein and DNA shown in space-fill mode. Yellow lines accentuate proximity to IN β4-α2 connector. (D) Q148H/G140S active site bound to BIC. δ+ indicates increased electropositivity of the His148 Nε2 proton. (E) The extended hydrogen bond network that couples Thr138 to His148 in the Q148H/G140S SIVrcm intasome. Black arrows indicate hydrogen bond donation; the corresponding interatomic distances are given in Ångstroms. Cryo-EM map of the same region is shown in fig. S20. (F) Long-range interactions of Ile74 and Thr97 with the metal chelating cluster via Phe121. Key amino acid residues are shown as sticks and semi-transparent van der Waals surfaces. Contacts between side chain atoms are indicated by double-headed dotted arrows with distances given in Ångstroms. Cryo-EM map showing definition of the side chain rotamers and the effects of I74M/T97A substitutions on the Phe121 side chain are shown in fig. S21.