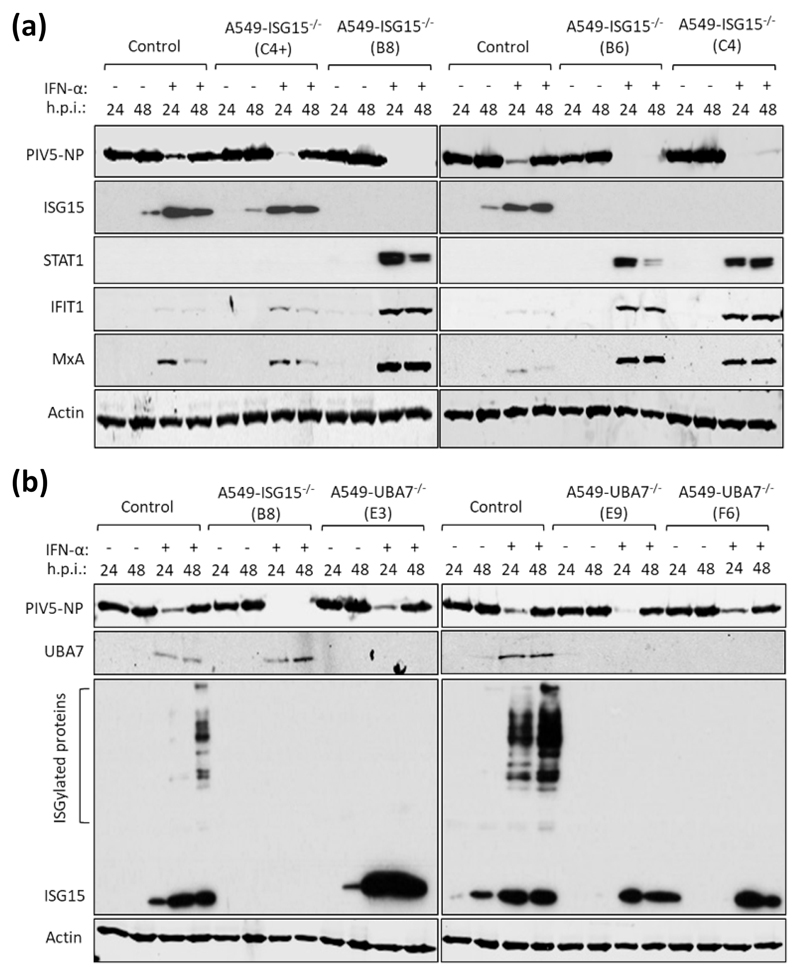
Fig. 3. IFN-α pre-treatment of ISG15-deficient cells leads to virus resistance which is independent of ISGylation.
(a) Control (naïve A549) and 4 independent clones of A549-ISG15-/- cells generated by CRISPR/Cas9 genome editing were treated with 1000 IU/ml IFN-α for 16 h or left untreated and then infected with PIV5 strain W3 (MOI = 10). Cells were harvested at 24 h and 48 h p.i. and processed for immunoblot analysis using antibodies specific for PIV5 nucleoprotein (NP), ISG15, STAT1, IFIT1, MxA and β-Actin. This experiment was independently performed twice. (b) UBA7 knockout cells were generated using CRISPR/Cas9 genome editing; Cas9-expressing A549 cells were first generated (following transduction with lentiCas9-Blast) and then transduced with lentiGuide-Puro expressing a single guide RNA that targeted exon 3 of the UBA7 gene. Knockout cells were single cell cloned and three were selected for further analysis. These cells were treated with IFN-α or left untreated, infected and processed as in (a) using antibodies specific for PIV5 NP, ISG15, UBA7 and β-Actin. This experiment was independently performed twice.