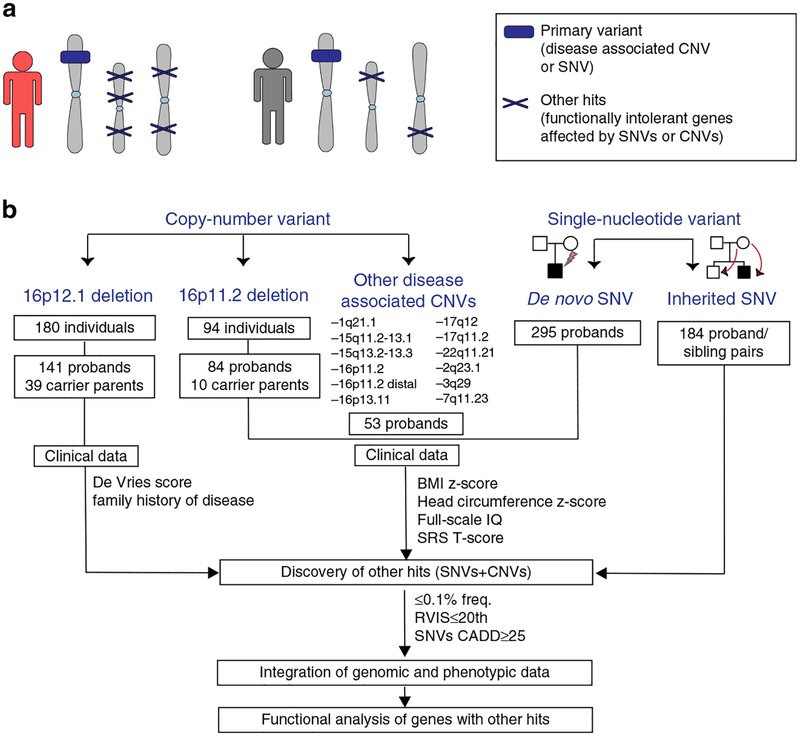
Fig. 1. Strategy for understanding the role of the genetic background in phenotypic variability of neurodevelopmental disease.
(a) Schematic of primary variants and other hits used in this study. Disease-associated variants shared among different individuals were considered as “primary variants” (blue box), and rare likely deleterious single-nucleotide variants (SNVs) or copy-number variants (CNVs) affecting functionally intolerant genes were defined as “other hits” (blue Xs). Individuals with a higher burden of other hits (in red) exhibit a more severe clinical manifestation compared with those carrying the same primary variant but with a lower number of other hits (in gray). (b) Combined clinical and genomic analysis of 757 probands and 233 family members carrying primary disease-associated variants (16p12.1 deletion, 16p11.2 deletion, 16 rare CNVs, de novo pathogenic variants in autism simplex cases, and inherited pathogenic variants in disease-associated genes) was performed to understand the role of rare (≤0.1%) likely deleterious variants (SNVs with CADD ≥25 and CNVs) in functionally intolerant genes (Residual Variation Intolerance Score [RVIS] ≤20th percentile) toward the variable manifestation of neurodevelopmental disease. SRS Social Responsiveness Scale