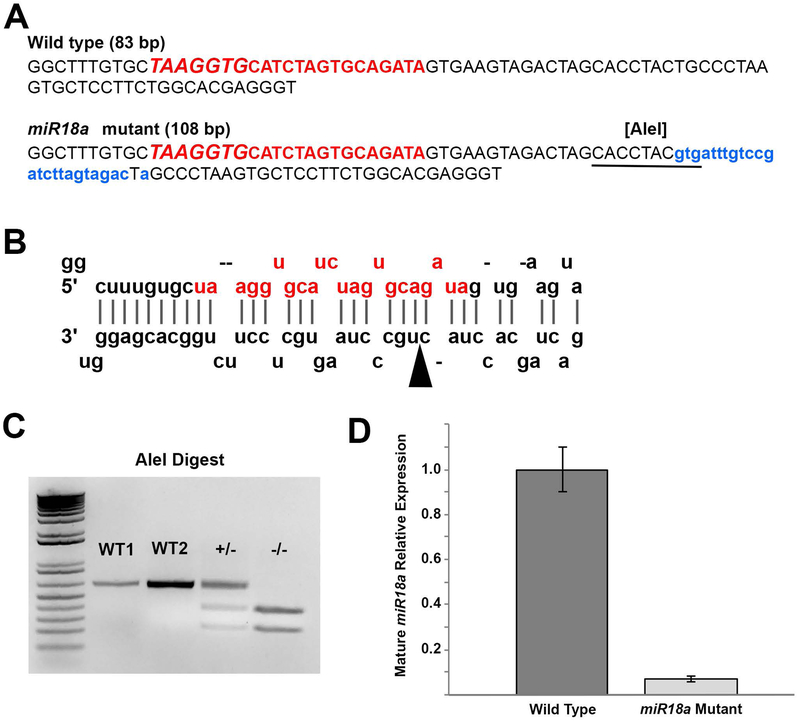
Figure 4. Characterization of the miR-18a−/− mutant line.
(A) Comparison between WT and mutant genomic sequences corresponding to pre-miR-18a and in the mutant, the 25 bp insertion is shown in lowercase, blue lettering and the introduced AleI restriction cut site is underlined. The uppercase red lettering shows the genomic sequence corresponding to mature miR-18a, with the seed sequence in larger italics. (B) The predicted stem loop arrangement for the WT pre-miR-18a RNA molecule with the mature miR-18a sequence shown in red (adapted from www.mirbase.org); in the mutant sequence, the 25-base insertion location is indicated by the arrowhead. (C) Genotyping of miR-18a mutants using AleI restriction digest. In WT fish, the 626 bp PCR product remains uncut, with clear intensity distinctions between 200 ng (W1) and 400 ng (WT2) PCR product. In heterozygous mutants (+/−) 50% (~200 ng) of the PCR product is cut into smaller fragments and in homozygous mutants (−/−) 100% (400 ng) of the PCR product is cut into smaller fragments. (D) TaqMan qPCR showing, compared with WT at 70 hpf, the relative absence of mature miR-18a in mutant fish. Error bars represent standard deviation on a single biological replicate of n=40 embryo heads per sample.