Figure 4.
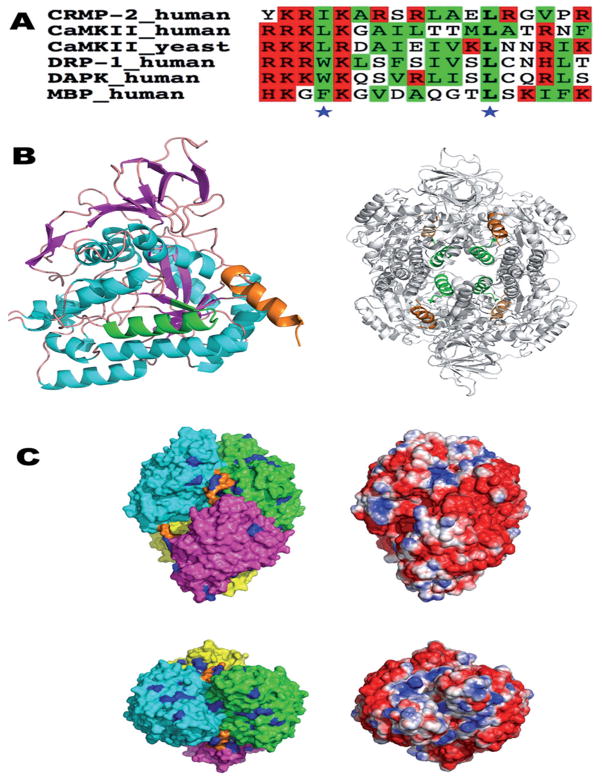
The putative CaM-binding site of CRMP-2 in the context of the 3D structure of CRMP-2. (A) A sequence alignment of the used CRMP-2 peptide with known CaM ligands. Basic and hydrophobic residues are highlighted in red and green, respectively, and the conserved hydrophobic positions in the 31–103 mode are indicated by asterisks. The sequences are from the CaM-binding domains of CaM-dependent protein kinase II (CaMKII), DAPK-related protein 1 (DRP-1), death-associated protein kinase (DAPK), and myelin basic protein (MBP). (B) The location of the CaM-binding domain (CaMBD) peptide sequence in the 3D structure of CRMP-2 (PDB entry 2GSE). On the left, only a monomer form is shown. The helices are in cyan, sheets in magenta, the first potential CaMBD (residues 258–275) in green, and the C-terminal CaMBD helix in orange. For the tetramer (right), the coloring of the CaMBDs is the same as in the left panel, while the rest of the tetramer is colored gray for clarity. The first CaMBD is located in the central lumen of the tetramer, while the C-terminal helices point outwards from the surface. (C) Surface representation of tetrameric CRMP-2. The top and bottom views are related by a rotation of 90°about the x axis. On the left, the molecular surface of the four monomers is color-coded according to polypeptide chain, and the C-terminal helix is indicated by orange. All arginine residues are colored blue. On the right, the solvent-accessible surface is colored based on electrostatics (red, −2 kT/e – blue, +2 kT/e), calculated using APBS.