Figure 1.
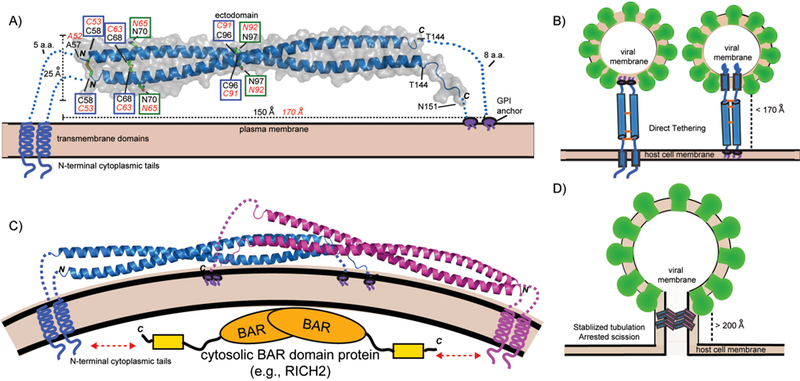
Structure of BST-2 and mechanisms of viral tethering. A: Crystal structure of mouse BST-2 ectodomain.[44] Structural features are highlighted: N-linked glycosylation sites = green boxes; disulfide bonds = blue boxes. Transmembrane domains, GPI anchors, and N-terminal cytoplasmic tails are shown as cartoons. Black labels correspond to mouse BST-2 sequence and features while the corresponding residues and values for human BST-2 are shown in red italics. B: The direct tethering models of BST-2 mediated inhibition of viral budding. Parallel BST-2 dimers incorporate one set of membrane anchors each into the viral membrane and host membrane. There is a 3–5 fold preference for incorporation of the GPI-anchor into the viral membrane (left) versus the other orientation (right).[51] C: A dimeric assembly created by crystallographic symmetry in the mouse BST-2 crystal structure. The dimer has a similar shape to BAR domains that bind membranes to generate or stabilize curvature. The N-terminal tail of BST-2 could bind cytoplasmic BAR domain proteins (e.g., RICH2)[55] to enhance mechanical stability in the stabilized tabulation model. (D): Stabilized tubulation and prevention of scission model of BST-2 mediated inhibition of viral budding. BAR-like BST-2 oligomers stabilize the tubulating membrane and prevent membrane scission. These tubules could be further stabilized by cytosolic BAR domain proteins that bind the cytoplasmic tail of BST-2 (e.g., RICH2)[55]. This hypothetical model is based on the observation of a BAR-like dimer in the mouse BST-2 ectodomain crystal structure [44] and the observation of long tube-like structures longer than the ectodomain of BST-2 (>200 Å) tethering HIV-1 VLPs to the plasma membrane by cryo-EM.[47]
