Figure 2.
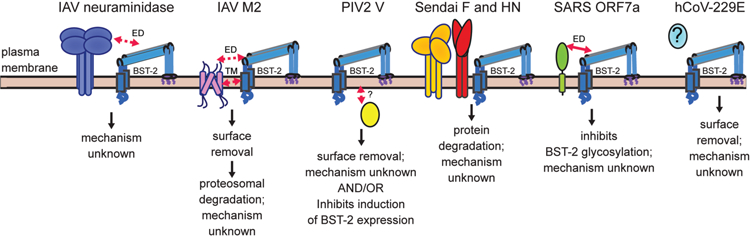
Respiratory virus antagonism of BST-2 and mode of action. Schematic summarizes what is currently understood about how various respiratory viruses antagonize BST-2. Structural domains demonstrated to mediate direct interactions are indicated by solid red arrows. Implied interactions are denoted by dashed red arrows. Antagonism of BST-2 by IAV is strain-dependent. In some strains, the neuraminidase protein partially inhibits IAV release.[18, 21, 33] This antagonism is thought to be mediated by the ectodomains (ED) of neuraminidase and BST-2. Another study suggests IAV antagonizes BST-2 using hemagglutinin, but the effect requires neuraminidase.[20] A more recent study suggests that the IAV M2 channel protein antagonizes BST-2 targeting interactions with the ectodomain and/or transmembrane (TM), causing surface removal.[34] The cytoplasmic V protein of PIV2 may antagonize BST-2 by two different mechanisms. It may bind and target BST-2 for surface removal [35, 80] and it may also inhibit induction of BST-2 expression.[65] How interaction is mediated is unknown. SeV F and HN proteins are required to degrade BST-2. However, it is unknown how interaction is mediated or the mechanism.[38] SARS CoV ORF7a ectodomain mediates an interaction with BST-2, which leads to inhibited glycosylation of BST-2 and its antagonism.[37] Human CoV-229E triggers surface removal of BST-2. The identity of the antagonist protein(s) is unknown.[36]
