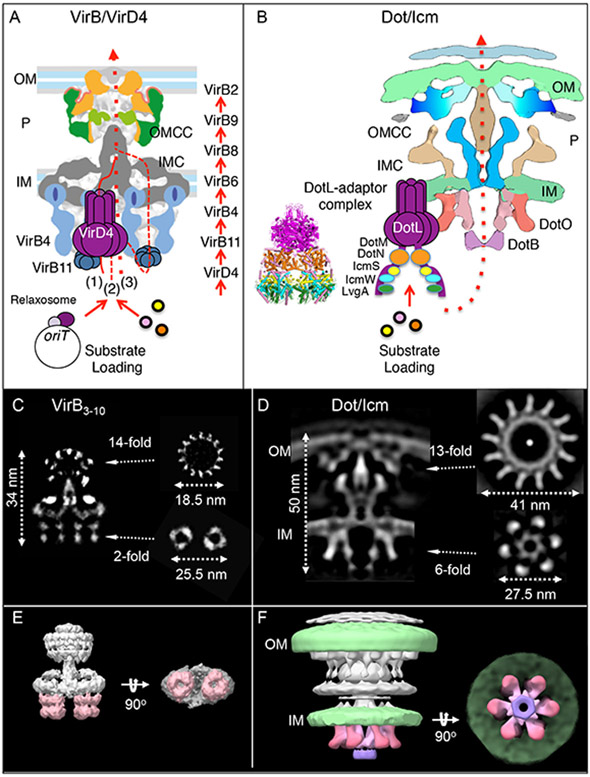
FIG. 2.
Architectures of the phylogenetically distant VirB/VirD4 and Dot/Icm T4SSs. A) A schematic of the VirB3-10 structure elaborated by the TrwR388 T4SS and solved by single-particle negative-stain electron microscopy (nsEM). A hexamer of the VirD4 receptor is fitted between the two hexameric barrels of the VirB4 ATPase. The VirD4 receptor recruits mobile genetic elements (MGEs) such as conjugative plasmids through recognition of components of the relaxosome (relaxase, accessory factors) assembled at the origin-of-transfer (oriT) sequence. VirD4 recruits protein substrates (colored dots) through direct or adaptor-mediated contacts. Substrates engage with the VirD4 receptor and are then delivered sequentially through a translocation channel composed of the VirB proteins listed at the right, as deduced from the TrIP assay (78). The route of transfer across the inner membrane is not known; substrates might be conveyed through the VirD4 hexamer (route 1, solid line), the VirB4 hexamer (2, small dashed line), or a channel composed of the VirB6 and VirB8 subunits (3, dotted line). Substrates then pass through the periplasm and across the outer membrane via an OMCC channel. B) A schematic of the L. pneumophila Dot/Icm T4SS solved by in situ cryoelectron tomography (CryoET) (16). The centrally stacked hexamers of the VirB4-like DotO and DotB form the cytoplasmic entrance to a channel that spans the entire cell envelope. The bell-shaped DotL-adaptor complex comprised of the hexameric nucleotide binding domain (purple) and C-terminal domain bound with DotN (brown), IcmS (yellow), IcmW (aqua), and LvgA (green); reprinted with permission by Kwak et al. (58). The DotL-adaptor receptor complex was not part of the visualized Dot/Icm T4SS (16,), and is provisionally positioned adjacent to the Dot/Icm T4SS. Upon loading of substrates, the DotL-adaptor complex is postulated to present effectors to the DotB/DotO energy center for delivery through the central channel. C-F) Comparison of the R388-encoded VirB3-10 substructure and the L. pneumophila Dot/Icm T4SS. C) A central section through the longitudinal plane of the VirB3-10 single-particle reconstruction with cross-sections of the OMCC and IMC at the positions indicated. D) A central section through longitudinal plane of a global average structure of L. pneumophila Dot/Icm T4SS with cross-sections at the positions indicated. E) A 3D surface rendering of the VirB3-10 substructure shown in side and bottom views. The side-by-side hexameric barrels of the VirB4 ATPase are colored pink. F) A 3D surface rendering of the Dot/Icm T4SS shown in side and bottom views. The bacterial membranes are in green and the DotO and DotB hexameric ATPases comprising the entrance to the translocation channel are in shaded pink and purple, respectively.