Abstract
Biologic treatment of type 1 diabetes (T1D) with agents including anti-CD3 (otelixizumab and teplizumab), anti-CD20 (rituximab), LFA3Ig (alafacept) and CTLA4Ig (abatacept) results in transient stabilization of insulin C-peptide, a surrogate for endogenous insulin secretion. With the goal of inducing more robust immune tolerance, we used systems biology approaches to elucidate mechanisms associated with C-peptide stabilization in clinical trial blood samples from new onset T1D subjects treated with the B cell depleting drug, rituximab. RNA sequencing (RNA-seq) analysis of whole blood samples from this trial revealed a transient increase in heterogeneous T cell populations, which were associated with decreased pharmacodynamic activity of rituximab, increased proliferative responses to islet antigens, and more rapid C-peptide loss. Our findings illustrate complexity in hematopoietic remodeling that accompanies B cell depletion by rituximab, which impacts and predicts therapeutic efficacy in T1D. Our data also suggest that a combination with therapy targeting CD4+T cells may be beneficial for patients who do not respond optimally to rituximab alone.
One Sentence Summary:
Elevated levels of CD4+ T cells predict poor response to rituximab in subjects with new onset type I diabetes.
Introduction
The therapeutic goal for type 1 diabetes (T1D) is to preserve β-cell function, which is commonly monitored by measuring insulin connecting peptide (C-peptide) levels as a proxy for islet function. Biologic therapies with distinct immunologic mechanisms of action, including anti-CD3 (otelixizumab and teplizumab), anti-CD20 (rituximab), and T cell costimulation blockade (abatacept), are partially effective in individuals newly diagnosed with T1D (1–7). Phase II studies with these agents have demonstrated transient stabilization of C-peptide levels in some individuals (non-progressors), but not others (progressors). However, in all cases, after a 6–12 month lag, both progressors and non-progressors lose C-peptide at the same rate as the placebo or control groups (2). Importantly, many untreated newly diagnosed individuals also show stabilization of C-peptide levels during the natural history of the disease post clinical diagnosis (8). The molecular basis for lack of immune tolerance, and heterogeneity in response to therapy and/or during natural progression is currently unknown.
To identify mechanisms involved in preservation of β cell function, we have devised combined systems biology and flow cytometry approaches with peripheral blood samples from clinical studies of treatment with biologic agents of newly diagnosed T1D subjects. In subjects treated with the humanized OKT3 monoclonal antibody (teplizumab) in the AbATE trial (4), we demonstrated accumulation of partially exhausted CD8+ T cells in the subset of patients showing the greatest stabilization of C-peptide levels (9). To investigate the generality of T cell exhaustion as a therapeutic mechanism in T1D, we applied the approaches we used with the AbATE study to the analysis of banked samples from a phase II trial of the anti-CD20 monoclonal antibody rituximab in new-onset T1D patients (5, 10). This study demonstrated a significant, but, transient, delay in loss of C-peptide production in treated subjects, with extensive heterogeneity in response (5, 10). In the present study, we have applied whole genome RNA-seq and flow cytometry analyses to identify molecular and cellular mechanisms associated with response of new onset T1D subjects to rituximab. Our goals were to identify unbiased signatures in whole blood that characterize progression versus non-progression following rituximab treatment; to compare and contrast these signatures with those seen in untreated subjects and subjects treated with teplizumab; and to determine how these signatures might suggest improved treatment for T1D.
Results
Whole blood RNA-seq analysis from T1D subjects treated with rituximab.
The rituximab trial was a randomized, double-blind study of patients (N = 87) with newly diagnosed T1D assigned to receive infusions of rituximab or placebo on days 1, 8, 15, and 22 of the study. The primary outcome was the geometric mean area under the curve (AUC) for the serum C-peptide level during the first 2 hours of a mixed-meal tolerance test assessed after 1 year. We obtained RNA-seq data from a subset of subjects in the original trial (N= 56 subjects originally), sampled at different visits (0, 26, 52, 78 and 104 weeks) for a total of 205 samples (mean ~4 samples per patient). To ensure against sample misidentification sometimes associated with large specimen collections (11), we compared reported sex with expression of X and Y chromosome genes (Methods). We also compared kinship coefficients derived from pairwise combinations of single nucleotide polymorphisms (SNPs) in RNA-seq reads from inter-and intra-subject libraries (Methods). Of 205 initial samples, 195 yielded high-quality RNA-seq data consistent with subject annotation, and were used in downstream analyses. Details of the study population and samples and how they compared with the original trial population are shown in Table 1. RNA samples were not available for 31/85 (~36%) of the subjects originally included in the trial. The subjects not included tended to be the younger subjects, who are more difficult to subject to blood sample collection. For example, the youngest subject for which RNA was available at 26 weeks was 13 years of age, whereas subjects as young as 8 years of age were included in the original study. Although the subset of subjects we analyzed by RNA-seq tended to be slightly older overall and have slower rates of progression relative to subjects in the full trial set, neither these or any other differences examined reached significance (p-value >0.05, Table 1).
Table 1.
Characteristics of the Study Groups.
| Previous study (19) | This study | ||||
|---|---|---|---|---|---|
| Variable | Rituximab | Placebo | Rituximab | Placebo | p-value 4 |
| No. subjects | 55 | 30 | 37 | 17 | NS |
| No. samples | NA | NA | 135 | 60 | NA |
| Gender | |||||
| M | 35 | 18 | 25 | 11 | NS |
| F | 20 | 12 | 12 | 6 | NS |
| Age 1 | 19.8 + 8.6 | 17.9 + 7.9 | 22.2 + 8.3 | 21+3 + 9.0 | NS |
| initial C-peptide 2 | 0.75 + 0.39 | 0.74 + 0.37 | 0.80 + 0.43 | 0.89 + 0.40 | NS |
| rate of C-peptide loss 3 | −0.46 + 0.45 | −0.69 + 0.66 | −0.38 + 0.33 | −0.63 + 0.62 | NS |
Shown is a comparison of subject characteristics in the previously described in the rituximab trial (19) with those analyzed in this study (i.e., those giving good quality RNA-seq libraries). Plus–minus values are means ±SD. NA, not applicable; NS, not significant (p-value > 0.05).
years;
nmol/L/minute;
log(nmol/L/min) / year;
t-test.
Rituximab treatment triggered transient decreases in B cell genes and increases in T cell genes.
To determine effects of rituximab treatment on gene expression in peripheral blood, we compared profiles from rituximab- and placebo-treated patients at different visits using gene set enrichment analysis (GSEA) (ref.12). For these analyses, we employed a modular or gene set approach which focused on pre-defined groups (modules) of coordinately expressed and annotated genes (9). To construct these modules, we created correlation matrices between levels of selected marker genes and all other genes across a collection of immune cell RNA-seq profiles and used the top most correlated genes as immune molecular modules (13).
When tested on rank ordered lists of genes expressed in rituximab- versus placebo-treated groups, none of these gene modules differed significantly in expression at the Week 0 (baseline) visit. This demonstrates that these treatment groups had broadly similar gene expression profiles prior to the start of treatment (Figure 1A). At the Weeks 26 and 52 visits, a set of gene modules (N = 4) were expressed at significantly lower levels in the rituximab-treated subjects. By week 78, expression of genes in these modules had returned to baseline levels (Figure 1A) and remained at baseline levels at week 104. In contrast, at the Week 26 visit, a separate set of gene modules (N = 21) was expressed at significantly higher levels in rituximab-treated subjects. Over-expression of genes in these modules also was transient, and their levels had returned to baseline by week 52 (Figure 1A). Individual module enrichment plots for CD19.mod (down-regulated), and GZMK.mod, CD2.mod, and CDH3.mod (up-regulated) genes are shown in Figure 1B. CD19.mod was chosen because it is representative of the four B cell modules; GZMK.mod, because it represented a T cell module that was not associated with disease progression; and CD2.mod, and CDH3.mod because they were associated with disease progression (see below).
Figure 1. Rituximab treatment triggered transient changes in whole blood gene module expression.
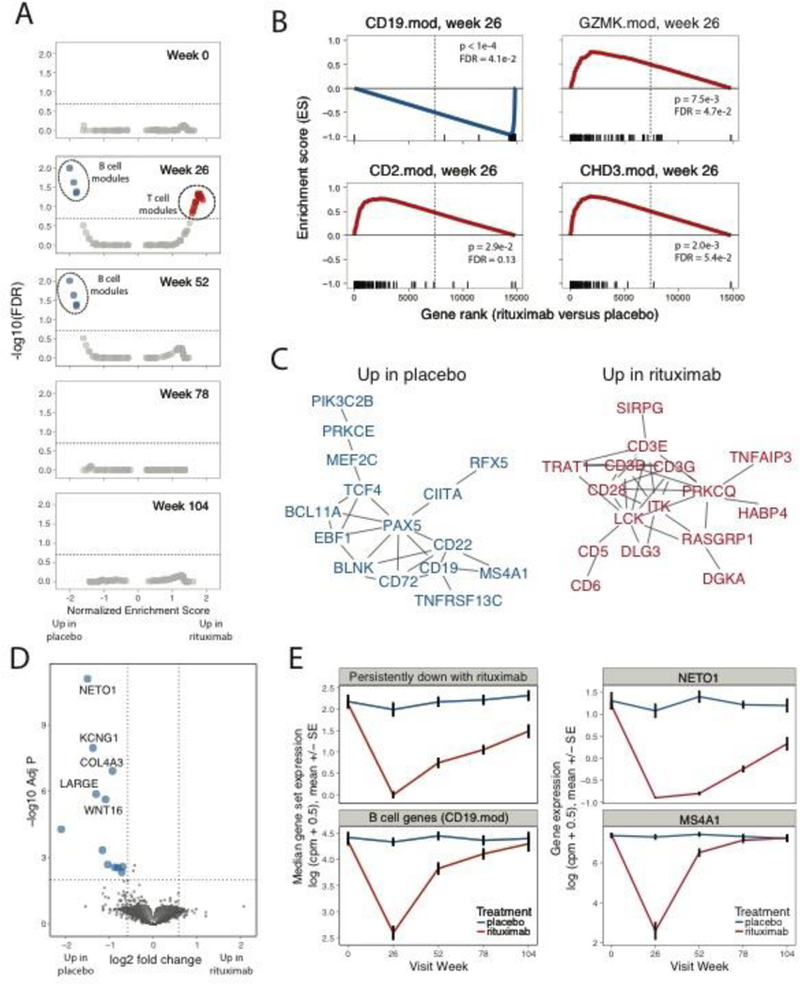
(A-B) GSEA (12) comparing rituximab- to placebo-treated patients. (A) Under/over-representation of specific gene sets in whole blood signatures from rituximab-treated individuals over the course of the study. Horizontal line indicates FDR of 0.20. Statistical tests were performed using the GSEA tool (http://software.broadinstitute.org/gsea/downloads.jsp). Blue, over-expressed in Placebo-treated subjects; Red, over-expression in rituximab-treated subjects. This analysis included 30, 28, 30, 27 and 20 rituximab-treated subjects at the 0, 26, 52, 78 and 104 week visits, respectively; and 13, 10, 12, 15 and 8 placebo-treated subjects at the same visits. (B) Selected gene sets (modules) significantly under/over-represented in rituximab-treated individuals at Week 26. X axis, enrichment score; Y axis, gene rank in rituximab- versus placebo-treated samples. Rug plots along the X-axes show differential expression ranks of module genes relative to all genes. (C) STRING network (14) of PPI interactions among genes in the leading edge of gene sets significantly up-regulated in rituximab-treated patients at Week 26. Shown are network graphs representing the unions of genes found multiple down- or up-regulated modules (>1 or >4, respectively). To minimize the size of the graph, vertices (genes) were filtered to have degrees (number of adjacent connections or edges) >1 and to represent vertices not farther than 3 connections from another fixed vertex (neighborhood). Vertices are colored as in Figure 1A. Vertex sizes are proportional to the rank of regulation shown in the rug plots in Figure 1B. D) Differential expression of genes between the placebo- and rituximab-treated patients at the 78 week visit, performed using limma-voom (17). Left, Horizontal dotted line represents FDR=0.01, vertical dotted lines represent fold change of +/− 1.5; Center, expression of module gene sets; Right, expression of representative individual genes over time in placebo-treated patients. Upper panels show genes persistently down-regulated with rituximab treatment, lower panels show B cell module genes (CD19.mod) and an established individual B cell marker gene, MS4A1 (CD20). There were N = 13 placebo and N = 30-rituximab-treated subjects tested at week 0, respectively; and N = 15 placebo and N = 27 rituximab treated subjects at week 78. Values are means across patients; error bars show +/− 1 standard error of the mean.
Closer examination of down-regulated gene modules revealed that they comprised highly overlapping B cell gene sets (Figure S1A). Genes from either the union of these gene sets (Figure 1C, Figure S1B) or from a selected individual module (Figure S2A) formed highly interconnected graphs when projected onto a Protein-Protein Interaction (PPI) network (14). The network graphs were enriched for interactions versus random sets of genes and contained well- known B cell genes (CD19, MS4A1 (CD20), etc.). Interconnected genes also were significantly enriched for annotations with the Gene Ontology (GO) (ref. 15) term for “B cell activation” (Figure S1B, Figure S2A). These findings demonstrate transient under-expression of B cell gene modules in whole blood following rituximab treatment, consistent with the depletion of B cells in peripheral blood seen clinically (5, 10).
We next examined the set of 21 up-regulated gene modules, which comprised five clusters of overlapping gene sets (clusters 1 and 3–6, Figure S1A). The union of genes from these up-regulated modules (Figure 1C, Figure S1B), or from selected individual modules (Figure S2A) also yielded highly interconnected PPI networks. Individual regulated genes included (Figure 1C, Figure S2A): T cell genes involved in variety T cell processes, including T cell receptor (CD3E, CD3D, LCK), and co-stimulation (CD28, ICOS); as well as markers for conventional (CD5), regulatory (Treg) (FOXP3, CTLA-4, and IL2RA) and cytotoxic (GZMA, GZMH, GZMK) T cells. Several of these T cell markers have expression patterns that delineate different T cell populations (i.e., CD2 is highly expressed on effector T cell lineages whereas CD3D and CD3E are expressed on all T cells, etc.). Interconnected genes also were enriched with the GO (15) or KEGG (16) terms, “T cell receptor signaling pathway” (Figure S1B). Taken together, these findings suggest a broad increase in T cells following rituximab treatment. The increase in T cells was not a generalized response to B cell depletion, as expression of most non-B, non-T module gene sets (N = 86), including sets representing other cell types (e.g., monocytes and neutrophils), were unaffected (Figure 1A).
To further investigate the type(s) of T cells contributing to the up-regulated signature, we identified a broader spectrum of individual gene expression differences (17) between 0 and 26 weeks samples from rituximab-treated subjects (Table S1). Genes down regulated after treatment included many B cell genes (Table S1). To identify up-regulated genes sharing functional interconnections, we projected the signature up-regulated after rituximab treatment onto a PPI network (Figure S2B). Consistent with the module analysis, we detected multiple T genes in the up-regulated signature (TRAT1, ITK, CD3G, CD6, LCP2, ITK, etc.). We also detected in the up-regulated signature: genes preferentially expressed in activated T cells (ICOS, SPN, CD40LG); genes involved in adhesion and migration (ITGA5, ITGA6 and LIMS1); and genes involved in chromatin methylation/demethylation (KDM3A, DNMT1, DNMT3) and chromatin remodeling (HDAC4, CHD3, SETD1B). These findings suggest that the up-regulated T cell signature contains genes from T cells that are activated and migratory, and are undergoing epigenetic re-modeling.
While module analysis suggested that expression of most genes had returned to baseline by week 78, comparisons of individual genes in samples from rituximab- versus placebo-treated subjects at later times revealed that expression of some individual genes remained depressed and did not return to baseline levels by week 104 (Figure 1D, Table S1). Most strongly down-regulated 13among these chronically down-regulated genes was NETO1, which modifies activity of kainate (glutamate) receptors (18) and regulates immunoglobulin production by human B cells (19). These results suggest that rituximab treatment may modify glutamine metabolism in B cells and have persistent effects on B cell gene expression and perhaps function up to 2 years following rituximab treatment.
Confirmation of changes in levels of cell subsets following rituximab treatment using flow cytometry.
To confirm dysregulation of cell subsets following rituximab treatment, we compared modular gene expression levels in rituximab-treated subjects with levels of B and T cell subsets determined by flow cytometry (20) (Figure 2 and Figures S3–S4). As expected, expression of genes in the selected B cell module, CD19.mod, showed strong positive correlations with the percentages of CD19+ B cells in the lymphocyte population (Figure 2A, Figures S3–S4). Gene expression measurements for CD19.mod had a broader dynamic range and extended below the limit of detection by flow cytometry, where many samples had essentially no detectable CD19+ cells (Figure S3, Figure S4). Expression of genes in CD19.mod showed significant negative correlations with the percentages of T cell subsets, especially CD3+ and CD4+ T cells (Figure S3, Figure 2A). In contrast, expression of genes in GZMK.mod correlated positively with the percentages of CD8+ T cells in the lymphocyte population, and genes in CD2.mod and CHD3.mod correlated positively with the percentages of CD3+ and CD4+ T cells (Figure S3, Figure 2A).
Figure 2. Flow cytometry also demonstrates over-expression of T cell genes in rituximab-versus placebo-treated subjects.
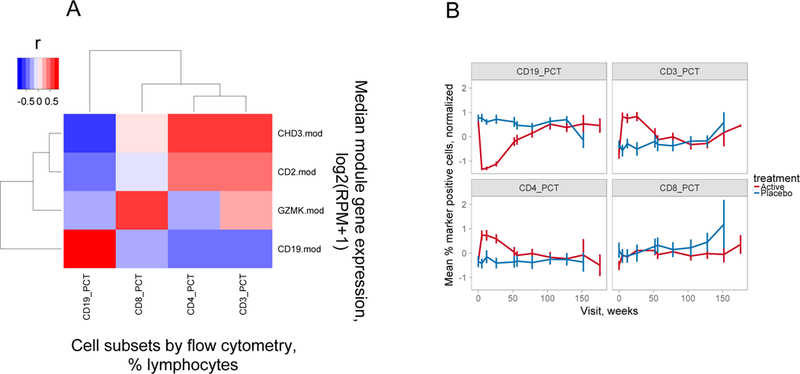
A) Correlation of modular gene expression with cell subset levels determined by flow cytometry. Shown is a heatmap representation of the correlation between modular gene expression measured by RNA-seq (Y axis) versus the percentages of cell subsets determined by flow cytometry (X axis). Gene expression was calculated as median log2 expression values in reads per million (RPM) +1 for all genes in the indicated module. Cell subsets were determined by antibody staining and were expressed as percentages of total lymphocytes (31). The magnitude of Pearson’s correlation coefficients (r) are represented by color intensity; Red, positive correlation; Blue, negative correlation. This plot was derived from 27 rituximab-treated subjects tested at week 26. B) CD3+ and CD4+, but not CD8+ T cell subsets were transiently overexpressed in rituximab-treated subjects. Percentages of the indicated cell subsets for all subjects at each visit were normalized by z-scores (value- mean of values)/SD of values). Shown are the z-score normalized mean percentages of the indicated cell subsets (+/− SD, Y axis) determined by flow cytometry (31) versus time of visit (X axis). There were 30–35 rituximab- versus 14–17 placebo-treated subjects tested at weeks 0–104 for each marker; and 25, 4 and 2 rituximab- versus 12, 2 and 1 placebo-treated subjects at weeks 128–176.
Importantly, correlations of module gene expression were stronger with lymphocyte populations calculated as proportions than absolute levels, suggesting that cell ratios altered by B cell depletion were important determinants of gene expression in whole blood. To further examine the cell differences detected using RNA-seq in Figure 1, we compared cell percentages of CD19+ B cells and CD3+, CD4+ and CD8+ T cells determined by flow cytometry in samples from both rituximab- and placebo-treated subjects across the course of the trial (Figure 2B). In this Figure, values were z-score normalized so as to facilitate comparisons between markers found at widely different levels. In agreement with Figure 1, the comparisons using flow cytometry showed the expected transient down regulation of CD19+ cells (5, 10), as well as significant transient up-regulation of CD3+ and CD4+, but not CD8+ T cells in treated subjects (Figure 2B, Figure S4). Taken together, these findings indicate that differential module gene expression after rituximab treatment was associated with decreased levels of B cells and up-regulation of CD3+ CD4+ T cells. Up-regulation of CD8 T cell genes was detected by RNA-seq, but levels of bulk CD8+ cells detected by flow cytometry did not differ between rituximab- and placebo-treated subjects (compare Figure 1 and Figure 2). Thus, differences in gene expression for GZMK.mod measured by RNA-seq were not reflected in bulk CD8+ cell levels measured by flow cytometry.
Quantifying rate of change in C-peptide loss in T1D patients.
A key question raised by the changes in lymphocyte populations described in the previous section is their relevance to response to therapy. To quantitate C-peptide loss throughout the course of the study, we constructed models for the rate of C-peptide loss, modifying the approach described by Pescovitz et. al. (10). The rate of C-peptide loss over time for both placebo- and rituximab-treated was highly variable (Figure S5A). However, when examined in log units, these curves were well-fit by linear models (Figure S5B), using patient as a random effect for slope and intercept, with a fixed slope effect by treatment group. These models captured a large percentage of the variation in C-peptide values both among patients and over time (pseudo-R2 = 0.94) (Figure S5C). Thus, loss of C-peptide is well-captured by a linear rate in log units (or exponential decay in absolute units), and resembles a first order decay reaction. Alternative models with additional variables, such as quadratic terms for change over time, did not substantially improve the fit.
The rate of change for rituximab-treated patients was greater than for placebo-treated subjects, though the difference did not reach significance (Figure S5D; p=0.066, consistent with results previously reported for C-peptide at the two-year visit (10). We also tested a half-life model of C-peptide loss, where the half-life is the time required for the C-peptide concentration to decrease to one-half its initial value (t½ = 0.693/k, where k is the rate of C-peptide decline in log units). We classified subjects as “progressors” if the half-life of C-peptide decline was less than the study period (104 weeks), and “non-progressors” if C-peptide half-life was longer than the study period. Samples classified as progressors by C-peptide half-life were reciprocally related to those designated previously as responders to treatment (20), with 13/17 non-responders versus 7/26 responders classified as progressors (p-value = 0.0020, Fisher’s test). We concluded that the half-lives of C-peptide decline were suitable metrics with which to investigate the effects of dysregulated T cell levels on T1D progression.
Differences in T cell gene module expression at Week 26 predict the rate of C-peptide decline in rituximab-treated patients.
Because T cell genes were significantly up-regulated in the rituximab-treated group after treatment, we hypothesized that the magnitude of T cell gene expression changes in the rituximab-treated patients may reflect underlying differences in the biological effects of treatment. To test this hypothesis, we utilized a previously described strategy (13) to test modular gene expression for the ability to predict patient progression after rituximab treatment. We first divided rituximab-treated subjects into two groups for each module, based on level of expression of module genes. We then compared progression to half-maximal levels of C-peptide in the two sets of patients using Kaplan-Meier (KM) analysis. To avoid extrapolation of the C-peptide data beyond the actual data points, we capped the maximum time to progression for each subject to the length of the study.
To determine appropriate criteria for grouping subjects based on module gene expression, we evaluated different stratification cuts for comparing gene expression levels. We found that comparing of samples falling in the top-quartile of gene expression (module high) versus those in the bottom three quartiles (module low) produced results consistent with observed progression status. As a test case for our methods, we used expression of T cell modules CD2.mod and CHD3.mod genes at 26 weeks to stratify rituximab-treated subjects into module high (top quartile) versus module low (bottom three quartiles) subsets. In both cases, all 7 subjects from the module high group were progressors, as compared with 7/21 samples from the module low group (p-value = 0.0058, Fisher’s test). These preliminary results suggested that high expression of CD2.mod and CHD3.mod T cell genes at 26 weeks was associated with more rapid C-peptide decline or progression, and poor response to rituximab therapy.
We then tested all differentially expressed immune molecule modules (Figure 1) for their ability to predict time to half-maximal decline of C-peptide and ranked them by multiple testing-corrected p-values (Table S2). Module high and module low subsets for B cell gene modules expression did not significantly differ in their time to half-maximal decline (Table S2), nor did they show significant differences using KM plots (Figure 3A). Likewise, module high and module low subsets for expression of most T cell gene modules, including the CD8+ T cell-associated module GZMK.mod, did not significantly differ in their time to half-maximal decline (Table S1), nor did they show significant differences using KM plots (Figure 3A). Similarly, we showed that, module high and module low subsets representing monocytes and neutrophils (CD14.mod and CXCR1.mod, respectively) also did not significantly differ in their time to half-maximal decline (Table S2). In contrast, module high and module low subsets for several T cell modules showed significant differences even after multiple testing correction (FDR<0.1), including CHD3.mod, CD2.mod and CD28.mod (Table S2). For each of these T cell modules, the module high subsets were associated with more rapid loss of C-peptide, as shown for CD2.mod and CHD3.mod stratification in Figure 3A (FDR = 3e-3 for both). Qualitatively similar results were obtained when using the Responder/non-Responder designations used in the original studies (data not shown, FDR = 0.067 for both CD2.mod and CHD3.mod) (ref. 20). Taken together, the statistical evidence supports the conclusion that there was non-random association of high expression of T cells genes at 26 weeks with poor response (more rapid C-peptide loss) of some patients after rituximab treatment.
Figure 3. Expression of CD4+ T cell-, but not CD8+ or B cell-module-associated genes at 26 weeks predicts C-peptide loss in rituximab-treated patients.
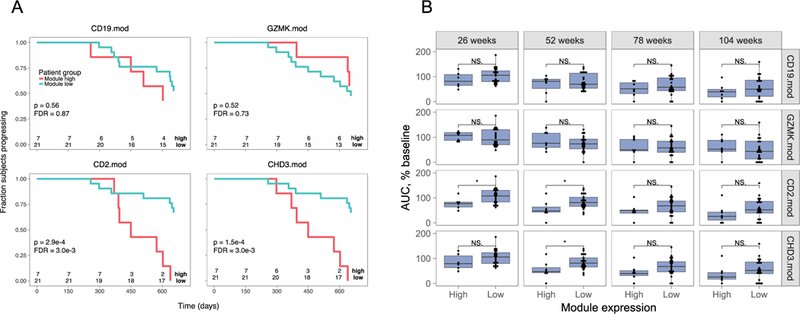
A) Rituximab-treated subjects were split into module high (top 25%) and module low (bottom 75%) groups based on log2 median module gene counts+1 values from RNA-seq profiles collected at week 26. Shown are Kaplan-Meier plots for progression, measured as time to 50% of baseline C-peptide, versus time. Survdiff (43) p-values were calculated and adjusted for multiple testing (40). Rituximab-treated subjects were stratified by median gene expression in B cell module, CD19.mod; CD8+ T cell-associated module, GZMK.mod; and CD4+ T cell-associated modules CD2.mod and CHD3.mod. Numbers at bottoms of panels are numbers of subjects at risk. Numbers of subjects tested are indicated at the bottom of each panel. B) Expression of CD4+ T cell-, but not CD8+ or B cell-module-associated genes at 26 weeks predicts C-peptide AUC levels at 52 weeks in rituximab-treated subjects. Shown are C-peptide AUC levels (% of baseline), across all visits of rituximab-treated subjects. Subjects were stratified into module high and nodule low subsets based on the expression of the indicated module gene expression at week 26, as described in Figure 3. Asterisks indicate level of significance of Wilcox test p-values: *, p-value<0.05 and p-value >=0.01. Numbers of subjects tested were the same as in panel A.
We conducted additional tests of the relationship of T cell module gene expression versus rate of progression of rituximab-treated subjects. We compared module gene expression in treated subjects at 26 weeks with C-peptide area under the curve (AUC) at other visits (Figure 3B). There was no significant difference in AUC values at any visit for comparisons of module high and module low subsets from CD19.mod nor GZMK.mod stratified subjects. In contrast, both CD2.mod and CHD3.mod stratified subjects showed a trend towards lower AUC values in module high subjects (evaluated at 26 weeks) in subsequent visits at 52, 78 and 104 weeks, with the differences at 52 weeks reaching significance (p-value <0.05) (Figure 3B). Importantly, previous studies showed similar significance between group level comparisons of rituximab- and placebo-treated subjects at 26 weeks, which was lost at 104 weeks (10).
To gain additional insight into the significance of our findings, we compared the predictive ability of T cell module gene expression with clinical parameters when considering both rituximab- and placebo- treated subjects. Stratification with both CD2.mod and CHD3.mod showed less significant differences when considering all subjects than when considering treated subjects only, likely because highly elevated T cell gene expression was not observed in placebo-treated subjects CHD3.mod expression (p-value = 0.047, Wilcoxon test, comparing expression in rituximab- versus placebo-treated subjects). Moreover, of the 21 subjects classified as progressors, 9/15 rituximab-treated progressors were also CHD3.mod module high, as compared with 0/6 placebo-treated progressors that were module high (p-value = 0.019, Fisher’s test).
We tested CD2.mod and CHD3.mod gene expression at 26 weeks alone and in combination with other variables using a Cox proportional hazards model. Although age (21) and rituximab treatment (5, 10) are known predictors of the rate of T1D progression, with this dataset both variables had less predictive ability at 26 weeks than the module high and low subsets determined by CD2.mod or CHD3.mod gene expression (p-values >0.05 for age and treatment, versus p-values = 0.01 for the module sets). Considering all three variables together did not greatly improve the predictive ability of either module alone.
Relationship of T cell module gene expression to pharmacodynamic and mechanistic parameters from the rituximab study.
We hypothesized that differential pharmacodynamic activity of rituximab might correlate with T cell dysregulation at 26 weeks. To test this possibility, we took advantage of the fact that some of the subjects in the rituximab trial were immunized with bacteriophage phiX174 (ref. 22), a de novo antigen which triggers a robust antibody (Ab) response. Rituximab treatment potently blocked primary and secondary Ab responses to phiX174 given 3 and 9 weeks after rituximab dosing (22), thereby demonstrating pharmacodynamic activity of the therapeutic agent. One year after rituximab dosing, tertiary and quaternary responses to phiX174 returned to ranges seen with placebo-treated subjects. When subjects immunized with phiX174 were stratified by CHD3.mod gene expression at 26 weeks, the module high subset showed a trend for increased Ab responses following primary and secondary immunizations shortly after rituximab treatment, but not following tertiary and quaternary immunizations one year after treatment (Figure S6, Figure 4A). Differences between the module high and module low groups were most pronounced during the secondary immunization (Figure 4A; repeated-measures mixed effect ANOVA, p-value = 0.034). It is important to point out that the power of these comparisons was limited by subject number, as only a fraction of the total subjects were both immunized with phiX174 and available for RNA-seq analysis (22) (only 26/37 (~70%) of rituximab-treated subjects available for RNA-seq were immunized). These increased Ab responses demonstrate reduced pharmacodynamic activity of rituximab in the subset of subjects with high CHD3.mod gene expression.
Figure 4. Relationship of T cell module gene expression to pharmacodynamic and mechanistic parameters.
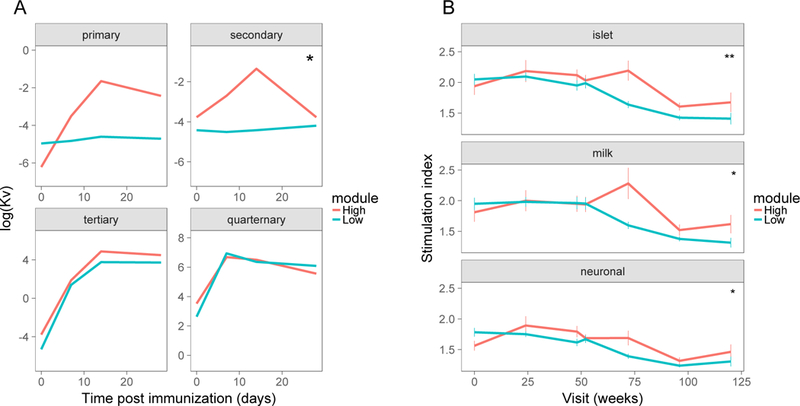
We stratified subjects into module high and nodule low subsets based on module CHD3.mod gene expression at week 26 and compared subsets for pharmacodynamic and mechanistic markers. A) Pharmacodynamic relationship of median CHD3.mod gene expression to phiX174 antibody responses following primary, secondary, tertiary and quaternary immunizations (22). Thin lines, individual responses; thick lines, mean responses. Asterisks indicate level of significance for p-values calculated from repeated measures ANOVA tests for module set as a fixed effect: *, p-value<0.05 and p-value >=0.01. This plot represents 14 rituximab-treated subjects (6 module high and 8 module low). B) Relationship of median CHD3.mod module gene expression to T cell proliferative responses to different antigens. Responses to multiple individual antigenic peptides were grouped thematically into islet, milk and neuronal antigens (20) and are presented as mean responses +/− standard error (SE). There were N = 6, 5, 6, 6, 6, 6, 4 module high and N = 12, 18, 19, 19, 17, 19, 7 module low subjects tested at the 0, 42, 84, 91, 126, 168, and 210 week visits, for N = 4, 5, and 4 islet, milk and neuronal peptides, respectively. Asterisks indicate level of significance for p-values calculated from repeated measures ANOVA tests for module set as a fixed effect: *, p-value<0.05 and **, p-value < 0.01.
We also hypothesized that increased activity of islet antigen-reactive T cells after rituximab treatment might result from T cell dysregulation at 26 weeks, and provide a potential mechanism for poor therapeutic activity. To test this possibility, we used a previously described data set that examined T cell proliferative responses to diabetes-associated and other antigens in peripheral blood T cells of subjects from the rituximab trial (20). When stratified by CHD3.mod gene expression at 26 weeks, the module high subset showed increased proliferative responses to islet, milk and neuronal antigens (Figure 4B), with greatest differences at 72 weeks. These trends were assessed using a repeated-measures mixed effect ANOVA model, which indicated significant elevations of responses to islet, milk and neuronal peptides in subjects with high CHD3.mod gene expression (p-values, 1.1e-3, 2.7e-2 and 1.1e-2, respectively, for module high versus module low comparisons). These findings support expansion of T cells with towards a broad range of antigenic specificities, including islet antigens, after rituximab treatment.
Discussion
Using modular gene expression approaches, we identified transient signatures in rituximab-treated individuals, with relative loss of B cells at 26 and 52 weeks. Although expression of a few genes likely related to B cells was persistently depressed, there were relatively minimal long-lasting lasting transcriptome alterations in whole blood resulting from a single course of rituximab therapy. We also identified a relative increase of T cell genes at 26 weeks, which was associated with CD3+, CD4+ and, to a lesser extent, CD8+ populations and returned to baseline levels by weeks 78 and 104, and predicted more rapid progression of T1D.While age also has been associated with variation in the rate of disease progression in T1D (21), presumably reflecting biological pathways that underlie disease heterogeneity, we found that, in this cohort, module gene expression more strongly predicted C-peptide loss than age. Furthermore, adding age to the model did not improve prediction.
Conclusions from our studies were necessarily limited in power by the size of the rituximab clinical study, and require validation in larger independent studies. Despite these caveats, our findings suggest that the increase in T cell gene levels following rituximab treatment provides a biomarker(s) for the efficacy of therapy. Our data also suggest a plausible mechanistic explanation for this finding: subjects with high T cell gene expression after treatment have increased relative numbers of islet antigen-reactive T cells following B cell depletion, which have the potential to trigger more autoimmune destruction of islets and more rapid loss of C-peptide.
A primary goal of these studies was to identify molecular and/or cellular signatures in whole blood of T1D subjects that were associated with response to rituximab therapy, and to determine whether these signatures are unique or treatment-specific (9). In a previous study, we saw a persistently increased whole blood signature of partially exhausted CD8+ T cells in patients having a favorable response to teplizumab (9). In this present study, we saw elevations in expression of T cell genes at week 26 (Figure 1A–B), but these elevations were associated with poor, not favorable response. Moreover, genes associated with response to therapy were better correlated with CD4+ than CD8+ T cells. Instead, the T cell responses we observed at 52 weeks therefore suggest a more generalized T cell response than what we observed with teplizumab-treated subjects. As different subjects were involved in the teplizumab and rituximab trials, it is not known how progressor and non-progressor subsets would have fared when treated with the other agent.
Another goal of our studies was to determine how treatment-specific signatures compared to signatures occurring during natural progression of untreated individuals. In this study, placebo-treated subjects had lower overall CHD3.mod expression. While numbers of subjects were small, they suggest that placebo-treated progressors did not have T cells elevated to the same extent as rituximab-treated subjects. Future studies using larger data sets will be required to provide additional insight into heterogeneity of mechanisms involved in progression of untreated T1D subjects. Taken together, however, our results from this study show that despite the similar clinical responses following treatment with teplizumab and rituximab (2), whole blood signatures were treatment-specific.
The relative gain in T cells after rituximab therapy was consistent with a previous study showing increased frequencies of islet antigen- reactive and other populations of antigen-specific T cells after rituximab therapy (20). Using the same data set, we showed that subjects stratified by expression of genes comprising a T cell module (CHD3.mod) also show increased proliferative responses to several distinct antigenic themes. Our results also show differences in levels of bulk CD3+ and CD4+ T cell populations, which are present at much higher frequency than rare antigen-specific cells. Our gene expression results also suggest an increase in CD8+ T cells, though this was not verified by flow cytometry, suggesting that the gene expression measurements may be more sensitive, or that they detect a subpopulation of CD8+ cells not measured by flow cytometry.
Taken together, our current results differ in several ways from the earlier study (20). Surprisingly, the previous study showed that elevated proliferative responses to islet antigens at 6–12 months, and which seemingly should be associated with worse outcome, were associated with beneficial short-term changes in the AUC of the C-peptide response (baseline to 6 months) (20). However, the relationship of these proliferative responses to AUC C-peptide at 1 and 2 years was not previously examined. Indeed, our data demonstrate that elevation of a broad-based spectrum of T cell genes at 6 months predicted faster progression (worse outcome) longer term. Moreover, we show that islet and other antigen responses are linked to a much broader based elevation of T cell levels and more rapid disease progression, following rituximab therapy than previously suspected.
An important question regarding the increase in T cell signatures following B cell depletion is whether this represents a specific process or whether it happens by default as the T cell compartment comprises a larger proportion of the cells isolated from peripheral blood. Although conclusions on individual T cell types may be confounded by blending of their profiles into bulk RNA-seq profiles, several lines of evidence suggest that the T cell signatures are specific. First, the set of gene modules tested contained modules representing other cell types that were not detected as changing (monocytes, neutrophils). In addition, not all T cells were elevated by flow cytometry (e.g., CD8+ T cell levels did not differ before and after treatment). We also note that our modular signatures (Figure 1C) contained numerous genes elevated on T cell activation (CTLA-4, FOXP3, IL7R, KLRG1, etc.). Finally, individual gene signatures suggested specificity in that the T cells that accumulated were associated with genes involved in T cell activation, migration, and epigenetic re-modeling. Together with previous studies on increased T cell proliferative responses (20), our data suggest that recovery of the hematopoietic compartment following B cell depletion with rituximab may lead to undesired expansion and/or increased activity of T cells.
Confirmation of the cell types, pathways and genes identified in our studies would require additional studies subject to sample availability. Such studies might initially involve single cell studies using expanded multi-parameter cytometry or single cell RNA-seq to better identify populations that accumulate after treatment. These single cell studies could then be followed by bulk RNA-seq studies with purified cell populations to compare accumulating cells with cells present in untreated individuals.
Most of the regulated T cell gene modules we observed were named for (i.e., associated with) (ref. 13) well-known T cell genes, with the exception of CHD3.mod. Although this module contained genes associated with T cell co-stimulation (Figure 1C), CHD3 itself is an autoantigen in some subjects with the inflammatory disorder, dermatomyositis (25), and functions as a chromatin-remodeling protein (26). CHD3-contining nucleosomal remodeling and deacetylase (NuRD) complexes cooperate with the zinc finger protein, Ikaros, to play important roles in T-cell development, including CD4 versus CD8 fate decisions and peripheral T-cell responses (27). Deletion of CHD3 (Mta2) causes abnormal T cell activation and lupus-like autoimmune disease in mice (27). Our findings implicate CHD3, and perhaps a role for chromatin remodeling, in T cells following rituximab treatment.
There are several possible explanations for poor outcomes of subjects having high T cell levels following rituximab therapy. First, differences in T cell levels following therapy may correspond to age-related differences in the role of B cells in disease. Rituximab’s beneficial effects in autoimmune disease may be explained in part by its interference with the ability of activated B cells to regulate T cell function. In T1D, rituximab had better efficacy in younger subjects (5), such that short-term efficacy that would have remained unnoticed if subjects only beyond 18 years of age had been recruited (7). As B cell levels are higher in younger subjects (28), the increased benefit of rituximab therapy in younger subjects may be related to elevated B cell levels and stimulation of T cell activation early in disease (i.e., in early epitope and antigen spreading and formation of memory T cells, etc.). This mechanism did not likely have a major impact in our study because we did not analyze RNA samples from the younger subjects having the greatest clinical benefit in the clinical study. Also arguing against this possibility is our finding that, in contrast to other cohorts, age was not a significant predictor of progression in the subjects we examined here. An alternative explanation is that rituximab given later in disease (or in older subjects) might have a converse impact by allowing increased numbers of pre-existing memory T cells to fill the immunological niche following B cell depletion. Whatever mechanism is involved, our studies suggest that the accumulating T cells were activated, migratory and actively undergoing epigenetic changes. Future studies including more balanced cohorts including younger subjects will be required to better elucidate any potential relationship between age, T cell levels and rate of progression in T1D.
Another possible explanation for poor outcomes with high T cell levels comes from clinical trials with systemic lupus erythematosus (SLE). In SLE, there have been suggestions of a feedback loop between rituximab infusion and increased levels of T follicular helper cells (23) that counteract efficacy of rituximab therapy (24). This mechanism has not been demonstrated in new onset T1D, but it parallels the association of poor response with high T cell levels reported here. Follow-on studies looking at levels of circulating Tfh cells following rituximab treatment might prove informative about whether this mechanisms is operative in T1D.
Yet another possible explanation for the observed variation in outcomes is non-uniform pharmacodynamic activity of rituximab across different subjects. Rituximab treatment suppressed de novo antibody production after phX174 immunization (22). Here, we confirmed these results and extended them by showing that subjects with high CHD3.mod gene expression had higher antibody titers than subjects with low CHD3.mod gene expression (Figure 4A, Figure S6). Thus, subjects with high T cells (high CHD3.mod gene expression) were less suppressed, indicating poorer pharmacodynamic activity of rituximab treatment in these subjects. While de novo antibody responses do not necessarily address immune mechanisms important for autoreactivity in T1D (i.e., epitope and antigen spreading, etc.), our results suggest that personalized dosing to maximize pharmacodynamic activity in individual subjects might improve efficacy of rituximab in T1D.
While T1D progression in clinical studies is commonly measured by C-peptide levels (29), there is no commonly accepted method to relate C-peptide levels to gene expression measurements throughout the course of a study. Clinical response to therapy (“Responders” versus “non-Responders”) is one possibility, but these designations have not been not made in a consistent manner between studies (4, 10, 22). For example, with the rituximab study, the Responder-non-Responder designation was based upon change in C-peptide AUC between baseline and the 6 month visit (20), whereas results from a single visit were used in the teplizumab study (4). Using results from a single visit or a difference between two visits, does not take advantage of the contributions of other visits to the overall patterns in the study and potentially introduces noise. The exponential decay model we describe here can be used to quantify the rate of C-peptide loss over time and in a consistent manner between studies, with less sensitivity to anomalous or missing values. Our new findings argue that exponential decay models described here and elsewhere (10) provide a new dimension to the study of disease progression and response to therapy.
Another potential translational implication of our findings is their support for biologic combination therapy. A single course of rituximab treatment in newly diagnosed T1D subjects leads to transient stabilization of β cell function, as measured by C-peptide levels, followed by a decline (5, 10). Similar findings have been seen following monotherapy with several other biologic agents (2). In organ transplantation, combination of biologic agents that work sub optimally as monotherapies resulted in long term graft acceptance (tolerance) (ref. 30). The transient nature of therapeutic effects in T1D suggests that combination therapies may be required to achieve long-lasting clinical benefit. Our studies show that an unintended consequence of B cell depletion by rituximab is increased number and/or activity of T cells that is associated with more rapid disease progression. This suggests that combination or sequential therapies of rituximab with other agent(s) that block T cell activity, such as teplizumab (4) or abatacept (3), might lead to more durable clinical effects.
Methods
Patient and sample selection.
All subjects were participants in the TrialNet phase II study of the effects of the anti-CD20 monoclonal antibody rituximab in new-onset T1D (TN-05) (ref. 5, 10). Subjects from the original trial were included in the current study if sufficient whole blood samples were available for RNA preparation (Table 1). All available high quality samples at regularly scheduled visits were utilized for each analysis. Numbers of subjects included in each analysis are indicated in the figure legends. TrialNet also provided clinical and flow cytometry data, collected as previously described (5, 10); phiX174 immunization results (22); T cell proliferation data collected in response to antigenic peptides (20); and 2-hour mixed-meal tolerance test (MMTT) results. C-peptide levels were calculated from MMTT results using the R package, flux, from the trapezoidal area under the curve (AUC) with measurements at 0, 15, 30, 60, 90, and 120 minutes.
Modeling rates of C-peptide change.
To estimate patient-level rates of C-peptide change over time, we estimated exponential decay using linear models fit to log-transformed C-peptide AUC measurements. We included patient-level random effects terms for the intercept and slopes, with treatment as a fixed effect. This allowed each patient to vary around a central value for each treatment group. We then extracted the patient-level coefficients from these models, and used the rate of change of log C-peptide over time as a measure of rate of T1D progression. This approach provided a single continuous measure of progression per patient, and allowed inclusion of patients even if data were missing from one or more visits. Finally, we used these models of C-peptide change over time to estimate each individual’s time to 50% of baseline C-peptide, as the half-life of the exponential decay, for use in downstream Kaplan-Meier survival analyses.
Whole blood RNA-seq pipeline analysis.
Samples of RNA purified from whole blood were obtained from TrialNet (https://www.trialnet.org/). Whole blood was collected in Tempus blood RNA tubes at the clinical site according to manufacturer’s instructions (Applied Biosystems, CA). RNA was isolated from whole blood samples using the Total RNA Isolation chemistry on an ABI Prism 6100 (Applied Biosystems, CA). Prior to sequencing, samples were globin-reduced with the GLOBINclear kit (Ambion, CA), and libraries were constructed from globin-reduced RNA using the Illumina TruSeq RNA Sample Preparation kit v2. Libraries were clustered on flow cells using the TruSeq Single Read Cluster Kit v3, followed by single-read sequencing for 50 cycles on a HiSeq2500 sequencer (Illumina, CA).
Base-calling was performed automatically by Illumina real time analysis software and demultiplexing was performed on Illumina BaseSpace after sequencing to generate FASTQ files; FASTQ reads were trimmed in a local Galaxy server in two steps: 1) hard-trimming to remove 1 3’-end base (FASTQ Trimmer tool, v.1.0.0); 2) quality trimming from both ends until minimum base quality for each read ≥ 30 (FASTQ Quality Trimmer tool, v.1.0.0) (ref. 31, 32). Reads were aligned in Galaxy using Bowtie and TopHat (TopHat for Illumina tool, v.1.5.0) (ref. 33). Read counts per Ensembl gene ID were estimated in Galaxy using htseq-count (htseq-count tool, v.0.4.1) (ref. 34). Sequencing, alignment, and quantitation metrics were obtained for FASTQ, BAM/SAM, and count files in Galaxy using FastQC, Picard, TopHat, Samtools, and htseq-count. Individuals conducting the RNA-seq laboratory and pipeline analyses were blinded to the sample descriptions. Data were deposited in the GEO repository under accession number GSE112594.
Code availability.
Data files and R code are available from the GitHub Repository (https://github.com/linsleyp/Linsley_Dufort_rituximab_TN-05).
RNA-seq sample identity checks.
We utilized two methods to ensure that proper sample identification was associated with RNA-seq profiles. Prediction of sex from RNA-seq data. RNA-seq reads were aligned to 2,539 loci mapping to the X chromosome and 589 loci mapping to the Y chromosome. From this alignment three metrics were calculated: the ratio of counts on the Y chromosome to counts on the X chromosome, the ratio of genes detected from the Y chromosome to genes detected from the X chromosome, and the ratio of squared counts on the Y chromosome to the total number of counts. Histograms of these three metrics were visually inspected to establish threshold values for predicting subject sex from each metric. The most frequent prediction from all three metrics was taken as the final predicted sex and compared to the reported sex of the subject. SNP-based kinship analysis of RNA-seq data. We used the mpileup utility (35)to call genetic variants from RNA-seq data; PLINK2 (ref. 36) to convert variant call files into the PLINK.bed format; and KING (37) to compute a kinship coefficient for pairwise combinations of RNA-seq libraries. Kinship coefficients computed on pairwise combinations of intrasubject libraries were then compared to the kinship coefficients for pairwise combinations of intersubject libraries.
RNA-seq data analysis.
We excluded samples with low quality RNA-seq data, defined as having fewer than 4 million total reads, less than 80% of reads aligned to the reference genome, or median coefficient of variation of read coverage greater than 1; 195 of 205 samples passed these quality filters. We normalized counts using the trimmed mean of M values (TMM) (ref. 38) as implemented in the BioConductor package edgeR (39). We included genes in analyses if they had greater than 1 count per million in at least two libraries. Differential expression of individual genes was determined with limma-voom (17). In addition to variables of interest, all models included patient sex as a covariate. Including age at diagnosis or cell counts from CBC differentials as covariates in the models did not significantly alter the results. Raw p-values were corrected for multiple testing using the Benjamini-Hochberg procedure (40). Gene set analyses were run with GSEA (12) on log-transformed, TMM-normalized counts. PPI interactions were obtained from STRING (14) (http://string-db.org/) or GeneMANIA (41) (http://genemania.org/) and visualized using Cytoscape (42) or the R package, igraph.
Statistical and graphical analyses.
Statistical tests were performed using the R programming language and software environment. Mixed-effects models were fit using the R packages, lme4 and lmer. C-peptide AUC values were modeled using the exponential decay model, log (C-peptide AUC) ~day + day:treatment + (1|id) + (day-1|id). Unless otherwise noted, statistical tests were two-sided. Variances were not assumed to be equivalent between different groups. For other tests using continuous, normally distributed variables, we used t tests; for non-normally distributed variables, Wilcoxon tests; for categorical variables, Fisher exact test; and for the probability of success in stratifying subjects by time of progression, the exact binomial tests. In all cases, we used well-established statistical tests with default settings. When parametric tests were used for plots, estimates of variation are provided. For comparison of phiX174 Ab levels and T cell proliferation over time, we utilized mixed effect repeated measures ANOVA models in R, using subject ID as a random effect (value~day*module + Error(id)); the reported p-values were for differences between module high and module low groups. The specific test used to derive each p-value is listed in the text. Kaplan-Meier survival analyses was performed using the R package, survival, and visualized using ggplot2 and the ggkm function (https://github.com/sachsmc/ggkm). Graphical visualizations were performed using R base graphics or the ggplot2 package.
Supplementary Material
Acknowledgments
We gratefully acknowledge the assistance and support of the TrialNet organization, especially Sarah Mueller, for providing samples, clinical and demographic data. We also acknowledge Jane Buckner, Jerry Nepom, Kevan Herold, Cate Speake and Hannah DeBerg for comments on the manuscript; Michael Dosch for the T cell proliferation data; Vivian Gersuk, Marty Timour, Kimberly O’Brien, and Quynh-Anh Nguyen for performing the RNA-seq analysis; and Michael Mason for performing the RNA-seq pipeline analysis. This work was supported by grants to PSL from the JDRF (1-PNF-2014–94-Q-R) and the NIH (DP3 DK104465–01), and by funding from the Immune Tolerance Network (ITN), NIH grant, 5UM1AI109565, awarded to Gerald T. Nepom. This work will support the mission of the ITN, which is to accelerate the clinical development of immune tolerance therapies. Samples for this study were obtain through an ancillary study to the TN-05 TrialNet Anti CD20 (Rituximab) Trial. TrialNet is currently funded by NIH grants U01 DK061010, U01 DK061034, U01 DK061042, U01 DK061058, U01 DK085461, U01 DK085465, U01 DK085466, U01 DK085476, U01 DK085499, U01 DK085509, U01 DK103180, U01 DK103153, U01 DK103266, U01 DK103282, U01 DK106984, U01 DK106994, U01 DK107013, U01 DK107014, UC4 DK106993, and the Juvenile Diabetes Research Foundation International (JDRF).
Footnotes
Conflict of Interest
The authors declare no competing interests.
References
- 1.Rigby MR, Ehlers MR. Targeted immune interventions for type 1 diabetes: not as easy as it looks! Curr Opin Endocrinol Diabetes Obes. 2014;21(4):271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenbaum CJ, Schatz DA, Haller MJ, Sanda S. Through the fog: recent clinical trials to preserve beta-cell function in type 1 diabetes. Diabetes. 2012;61(6):1323–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orban T, Bundy B, Becker DJ, DiMeglio LA, Gitelman SE, Goland R, et al. Co-stimulation modulation with abatacept in patients with recent-onset type 1 diabetes: a randomised, double-blind, placebo-controlled trial. Lancet. 2011;378(9789):412–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herold KC, Gitelman SE, Ehlers MR, Gottlieb PA, Greenbaum CJ, Hagopian W, et al. Teplizumab (anti-CD3 mAb) treatment preserves C-peptide responses in patients with new-onset type 1 diabetes in a randomized controlled trial: metabolic and immunologic features at baseline identify a subgroup of responders. Diabetes. 2013;62(11):3766–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pescovitz MD, Greenbaum CJ, Krause-Steinrauf H, Becker DJ, Gitelman SE, Goland R, et al. Rituximab, B-lymphocyte depletion, and preservation of beta-cell function. N Engl J Med. 2009;361(22):2143–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolb H, von Herrath M. Immunotherapy for Type 1 Diabetes: Why Do Current Protocols Not Halt the Underlying Disease Process? Cell Metab. 2017;25(2):233–41. [DOI] [PubMed] [Google Scholar]
- 7.von Herrath M, Peakman M, Roep B. Progress in immune-based therapies for type 1 diabetes. Clin Exp Immunol. 2013;172(2):186–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenbaum CJ, Beam CA, Boulware D, Gitelman SE, Gottlieb PA, Herold KC, et al. Fall in C-peptide during first 2 years from diagnosis: evidence of at least two distinct phases from composite Type 1 Diabetes TrialNet data. Diabetes. 2012;61(8):2066–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Long SA, Thorpe J, DeBerg HA, Gersuk V, Eddy J, Harris KM, et al. Partial exhaustion of CD8 T cells and clinical response to teplizumab in new-onset type 1 diabetes. Sci Immunol. 2016;1(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pescovitz MD, Greenbaum CJ, Bundy B, Becker DJ, Gitelman SE, Goland R, et al. B-lymphocyte depletion with rituximab and beta-cell function: two-year results. Diabetes Care. 2014;37(2):453–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodworth A PA. Chapter 4 - Sample Processing and Specimen Misidentification Issues In: Dasgupta A SJ, editor. Accurate Results Clin Lab [Internet]. On line: Elsevier; 2013. p. 35–51. [Google Scholar]
- 12.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Linsley PS, Speake C, Whalen E, Chaussabel D. Copy number loss of the interferon gene cluster in melanomas is linked to reduced T cell infiltrate and poor patient prognosis. PLoS One. 2014;9(10):e109760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tomita S, Castillo PE. Neto1 and Neto2: auxiliary subunits that determine key properties of native kainate receptors. J Physiol. 2012;590(10):2217–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sturgill JL, Mathews J, Scherle P, Conrad DH. Glutamate signaling through the kainate receptor enhances human immunoglobulin production. J Neuroimmunol. 2011;233(1–2):80–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herold KC, Pescovitz MD, McGee P, Krause-Steinrauf H, Spain LM, Bourcier K, et al. Increased T cell proliferative responses to islet antigens identify clinical responders to anti-CD20 monoclonal antibody (rituximab) therapy in type 1 diabetes. J Immunol. 2011;187(4):1998–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bollyky JB, Xu P, Butte AJ, Wilson DM, Beam CA, Greenbaum CJ, et al. Heterogeneity in recent-onset type 1 diabetes - a clinical trial perspective. Diabetes Metab Res Rev. 2015;31(6):588–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pescovitz MD, Torgerson TR, Ochs HD, Ocheltree E, McGee P, Krause-Steinrauf H, et al. Effect of rituximab on human in vivo antibody immune responses. J Allergy Clin Immunol. 2011;128(6):1295–302 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehrenstein MR, Wing C. The BAFFling effects of rituximab in lupus: danger ahead? Nat Rev Rheumatol. 2016;12(6):367–72. [DOI] [PubMed] [Google Scholar]
- 24.De Vita S, Quartuccio L, Salvin S, Picco L, Scott CA, Rupolo M, et al. Sequential therapy with belimumab followed by rituximab in Sjogren’s syndrome associated with B-cell lymphoproliferation and overexpression of BAFF: evidence for long-term efficacy. Clin Exp Rheumatol. 2014;32(4):490–4. [PubMed] [Google Scholar]
- 25.Ge Q, Nilasena DS, O’Brien CA, Frank MB, Targoff IN. Molecular analysis of a major antigenic region of the 240-kD protein of Mi-2 autoantigen. J Clin Invest. 1995;96(4):1730–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marfella CG, Imbalzano AN. The Chd family of chromatin remodelers. Mutat Res. 2007;618(1–2):30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu X, Kovalev GI, Chang H, Kallin E, Knudsen G, Xia L, et al. Inactivation of NuRD component Mta2 causes abnormal T cell activation and lupus-like autoimmune disease in mice. J Biol Chem. 2008;283(20):13825–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scholz JL, Diaz A, Riley RL, Cancro MP, Frasca D. A comparative review of aging and B cell function in mice and humans. Curr Opin Immunol. 2013;25(4):504–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludvigsson J C-peptide in diabetes diagnosis and therapy. Front Biosci (Elite Ed). 2013;5:214–23. [DOI] [PubMed] [Google Scholar]
- 30.Larsen CP, Elwood ET, Alexander DZ, Ritchie SC, Hendrix R, Tucker-Burden C, et al. Pillars article: long-term acceptance of skin and cardiac allografts after blocking CD40 and CD28 pathways. Nature. 1996. 381: 434–438. 1996. J Immunol. 2011;186(5):2693–7. [PubMed] [Google Scholar]
- 31.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15(10):1451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9(7):811–8. [DOI] [PubMed] [Google Scholar]
- 41.Montojo J, Zuberi K, Rodriguez H, Bader GD, Morris Q. GeneMANIA: Fast gene network construction and function prediction for Cytoscape. F1000Res. 2014;3:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Therneau T A Package for Survival Analysis in S_. R package version 2.37–4 2017. Available from: http://CRAN.R-project.org/package=survival.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


