Figure 2.
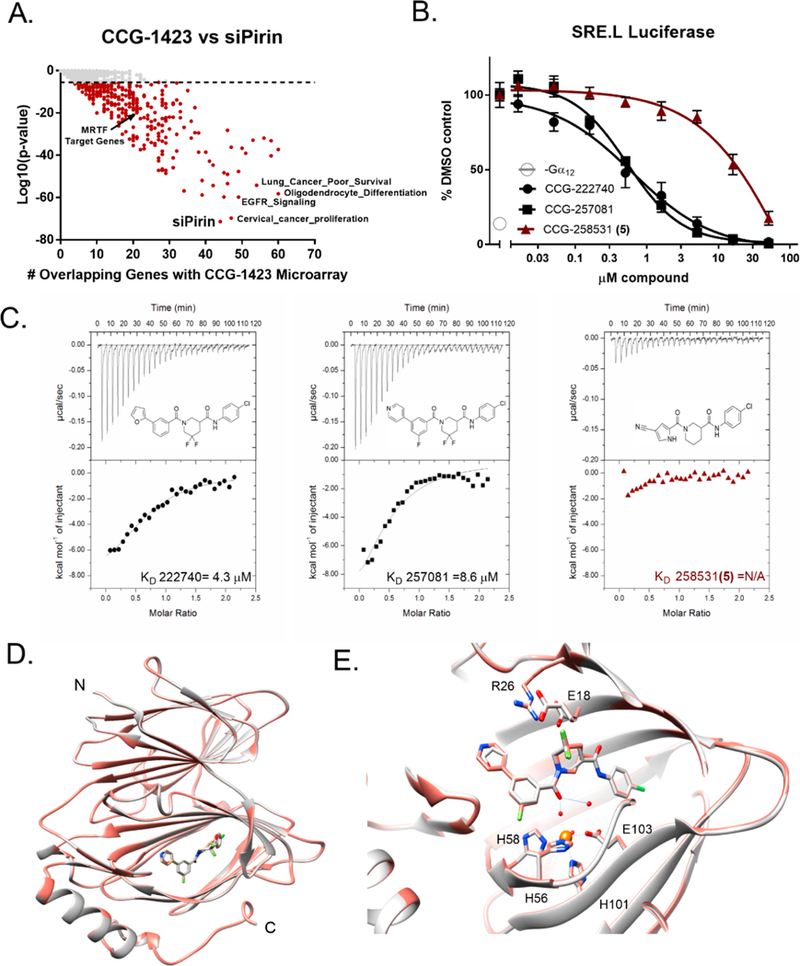
Pirin is a molecular target of CCG-222740 and CCG-257081. (A) Differential gene expression (based on fold change) was calculated for Pirin knockdown (GSE17551) and CCG-1423 treatment (GSE30188) and found to have high significance and an overlap of 44 genes in their data sets. (B) HEK293T cells cotransfected with Q231L Gα12 and a SRE.L luciferase reporter were treated with varying concentrations of CCG-222740, CCG-257081, or CCG-258531 (5). CCG-222740 and CCG-257081 inhibit Gα12 /Rho/MRTF mediated SRE.L luciferase much more potently than CCG-258531 (5). (C) Isotherm generated by ITC shows that CCG-222740 (left) and CCG-257081 (middle) have greater enthalpy changes upon binding to pirin, as compared to that of CCG-258531 (right), indicative of better binding to recombinant pirin. (D) Crystal structure overlap of pirin bound to CCG-222740 (gray), Protein Databank (PDB) code 6N0J, and CCG-257081 (salmon), PDB code 6N0K. (E) Detailed view of the compound binding pocket with overlaid CCG-222740 (gray) and CCG-257081 (salmon) pirin-bound structures. Also indicated is the metal ion (orange) as well as coordinating residues and water molecules (dashed lines), and hydrogen bonds (solid lines).
