Fig. 3.
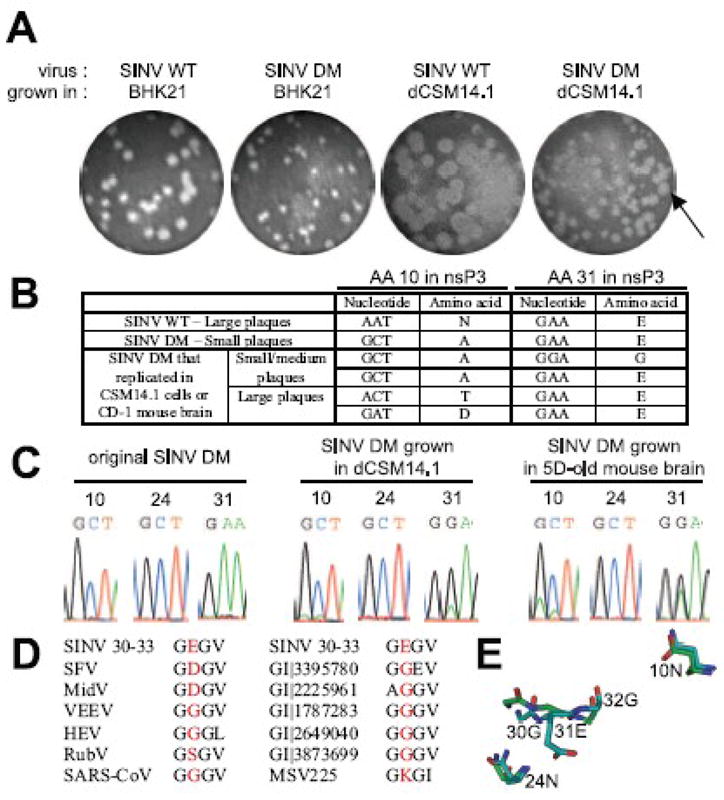
Reversions occur in the nsP3 macro domain of SINV DM during SINV replication. (A) The effect of mutations on plaque phenotype. P0 stocks of SINV WT and DM were produced in BHK21 cells, and the plaque phenotype observed (2 pictures on the left). 3 week-differentiated CSM14.1 (dCSM14.1) cells were infected with SINV WT or SINV DM, and viruses released into culture media were collected at 8 h post-infection. Plaque assay was performed in BHK21 cells to observe plaque phenotype (2 pictures on the right). (B) Analysis of plaque phenotype and reversions. Plaques from (A) were isolated, grown in BHK21 cells, and sequenced. (C)Assessment of reversions in SINV DM-infected CSM14.1 cells and the brains of CD-1 mice. P0 stock of SINV DM produced in BHK cells (left), virus released from SINV DM-infected differentiated CSM14.1 cells at 48 h post-infection (middle), and virus collected from the brains of SINV DM-infected 5 day-old CD-1 mice at 1 day post-infection (right) were sequenced. Electropherogram at amino acids 10, 24, and 31 of nsP3 is shown. (D) Alignment of SINV nsP3 amino acids 30–33 with other macro domain containing proteins. SFV, Semliki Forest virus; MidV, Middleburg virus; VEEV, Venezuelan equine encephalomyelitis virus; HEV, hepatitis E virus; RubV, rubella virus; SARS-CoV, severe acute respiratory syndrome coronavirus; GI|3395780, histone macroH2A1.1 from Gallus gallus; GI|2225961, Mycobacterium tuberculosis; GI|1787283, Escherichia coli; GI| 2649040, Archaeoglobus fulgidus; GI|3873699, Caenorhabditis elegans; MSV225, Melanoplus sanguinipes entomopoxvirus. (E) Homology modeling of the SINV nsP3 macro domain using Af1521 (a macro domain protein from Archaeoglobus fulgidus) as a template. The predicted structure of the nsP3 macro domain (blue) and the published structure of Af1521 (green) are superimposed, and close view image of nsP3 amino acids 10, 24, and 31 is presented.
