Abstract
The challenges in engineering bone scaffolds reflect the complexity of bone as an organ. The organic-inorganic hybrid system design aims to provide signals within a conductive apatite layer to promote cell adhesion, proliferation and ultimately differentiation into bone tissue. Dual functioning peptides designed to specifically adhere to the apatite layer, while promoting cell adhesion via cell recognition sequences, may increase cell adhesion, leading to increased osteogenesis. The aim of this study is to identify peptide sequences with preferential adsorption towards apatite-based materials. Bone-like mineral films and hydroxyapatite disks were panned with a phage library to elucidate peptide sequences with favorable adsorption. Peptide sequences were analyzed using the web-based biotechnology tool RELIC and validated with a modified ELISA, in addition to being investigated using a newly developed method of high-throughput computational modeling. Peptides having the highest affinity and greatest potential to be incorporated into a dual functioning peptide design are APWHLSSQYSRT, VTKHLN QISQSY and STLPIPHEFSRE. These experiments provide a method of rationally designing peptides that adhere to apatite and that may improve bone tissue regeneration. This work also provides structure for investigating peptide/protein adsorption on apatite substrates with varied carbonate, or other impurity, content.
Keywords: Phage display, Bone-like mineral, Peptide adhesion, Computational modeling
Introduction
Clinical challenges exist in treating orofacial and long bone defects caused by disease, trauma or congenital defects [Muschler and Midura, 2002; Wiesmann et al., 2004; Feinberg et al., 2005]. Traditional graft therapies used to treat bone defects often require a second surgical donor site and risk the chance of graft rejection [Yaszemski et al., 1996]. Bone tissue engineering approaches, developed to alleviate problems with traditional graft techniques, are designed to promote bone regeneration with the intent of eventual replacement of the biomaterial scaffolding with de novo tissue. Tissue engineering approaches to instructing bone growth include conduction, induction and/or cell transplantation [Alsberg et al., 2001].
Organic-inorganic hybrid scaffolds are designed to employ all 3 tissue engineering approaches of inducing bone formation. Deposition of bone-like mineral (BLM) onto degradable polymers provides cells a surface that is compositionally similar to bone. BLM can also adsorb and/or incorporate inductive biomolecules to provide spatiotemporally controlled biomolecular cues [Luong et al., 2006; Segvich et al., 2008], giving organic-inorganic hybrids the potential to influence cell fate. The challenge in designing an organic-inorganic hybrid construct is not only in fabricating a conductive synthetic apatite that cells prefer, but also in deciding which organic molecules to incorporate to induce and mimic nature’s ability to develop tissue that has the structure-function properties found in native bone.
Biomimetic methods aim to utilize cell recognition motifs by presenting extracellular matrix-derived sequences on implant surfaces [Garcia and Reyes, 2005]. Bioactive surfaces can initiate cell adhesion, which triggers a cascade of events that signal survival, differentiation and possibly enhanced osseointegration and bone repair [Damsky, 1999]. Dual functioning peptides designed to specifically adhere to BLM, while promoting cell adhesion via cell recognition sequences may provide an environment that increases cell adhesion. The aim of this study is to identify peptide sequences that preferentially bind to apatite-based substrates using phage display, and to validate the sequences with ELISA and computational molecular modeling. Phage display is a selection technique that presents approximately 109 unique 12-mer amino acid sequences on the protein coat of M13 phage. After multiple rounds of panning and amplification, this technique identifies DNA-encoded peptide sequences that have high physical affinity towards an identified substrate (for example, apatite).
Materials and Methods
Preparation of Biomimetic Films and Hydroxyapatite Disks
A 5% (w/v) 85:15 polylactic-co-glycolic acid (PLGA; Alkermes)-chloroform solution was cast on 12-mm-diameter glass slides and dried. The PLGA films were etched in 0.5 M NaOH for 7 min, rinsed in double distilled H2O and soaked in modified simulated body fluid (mSBF) for 5 days at 37° C [Luong et al., 2006]. The mSBF solution was prepared in double distilled H2O to include 141 mM NaCl, 4.0 mM KCl, 0.5 mM MgSO4, 1.0 mM MgCl2, 4.2 mM NaHCO3, 5.0 mM CaCl2 · 2H2O and 2.0 mM KH2PO4. mSBF, a supersaturated solution that enables self-assembly of a carbonated apatite layer onto the PLGA, was changed daily to maintain supersaturation and thermodynamic conditions conducive to heterogeneous nucleation. Hydroxyapatite (HA) disks (10 mm diameter or 4 mm thick) were pressed from powder (Plasma Biotal Ltd.) at 1 metric ton for 1 min and sintered at 1,350° C for 5 h. The disks were sonicated clean in 10 mM HCl, then in double distilled H2O. The HA disks and BLM films were soaked in double distilled H2O overnight prior to phage panning.
Phage Display and Qualitative ELISA on Apatite Materials
The Ph.D.12™ phage display library [New England Biolabs (NEB), manual No. E8110S], consisting of approximately 109 different phage with 12-mer amino acid linear peptide inserts, was panned against BLM films and HA disks. The NEB protocol was followed, using glycine/HCl, pH 2.2, to elute high-affinity phage. The phage experiment was repeated 3 times, with 3–4 rounds of panning per experiment. The DNA from a total of 243 clones was sequenced. A streptavidin control confirmed successful panning in each experiment. The publicly accessible web-based biotechnology tool REceptor LIgand Contacts (RELIC, http://relic.bio.anl.gov/index.aspx) was used to process high-affinity binding phage data acquired in the 3 phage display experiments [Mandava et al., 2004]. To verify selective binding to apatite-based substrates, 10 of the identified phage were analyzed using a modified ELISA on BLM films, HA disks, PLGA films and tissue culture polystyrene (TCPS) wells at 4 phage dilutions (106, 107, 108 and 1010 plaque-forming units). ELISA methods described in the NEB manual No. E8110S were followed. Briefly, each phage dilution was incubated on a substrate for 1.5 h with gentle rocking, and nonbinding phages were washed off with a Tris-based buffer. Horseradish peroxidase-conjugated anti-M13 antibody (GE Healthcare, No. 27-9421-01) was used to bind to adherent phage. Prior to detection, a 30% H2O2 (Corco, No. 1403)-2′,2′-azinobis(3-ethybenz-thiazoline-6-sulphonic acid diammonium salt (Sigma, No. A1888) solution in a 0.05 M citric acid solution, pH 4, was added to each well, and after a 50 min incubation at room temperature, an aliquot was read on a UV spectrophotometer (Bio-Rad SmartSpec 3000) at 410 nm. Background values from PLGA and TCPS were subtracted from the HA and BLM data.
Computational Modeling of Peptides on Apatite
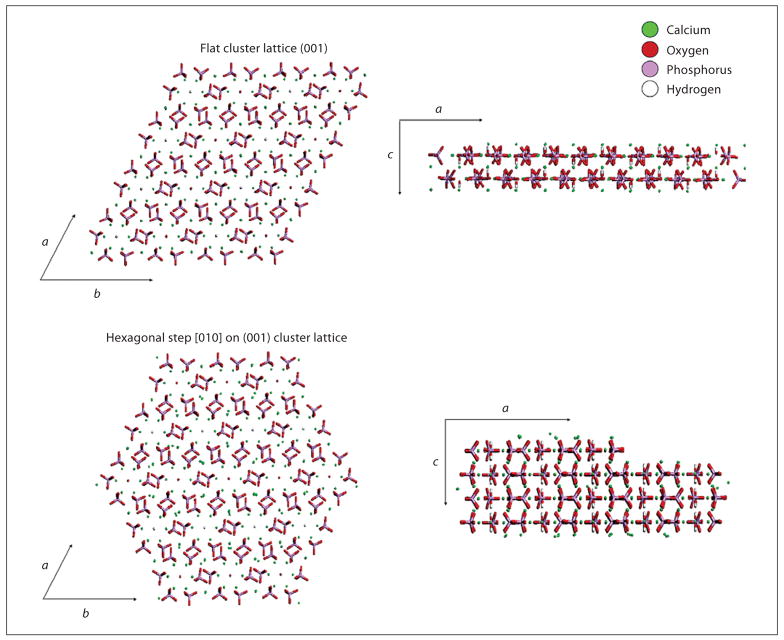
Using an HA force field developed from the Universal 1.02 force field [Rappe et al., 1992], peptide adsorption energies were calculated from simulations between HA and 19 peptides that emerged from multiple phage display experiments using molecular modeling techniques. Charge distribution within each peptide, calculated using a QEq charge equilibration [Rappe et al., 1991], was performed by distributing the appropriate charge of the peptide in a neutral environment, pH 7.2, across the molecule. After assigning charge, the individual atomic charges were kept constant in subsequent calculations to avoid energy fluctuation introduced during energy optimization of the structure of the isolated molecule and its adsorbate. To find the absolute energy minimum through optimization, molecular dynamics simulations under constant number of atoms, volume and energy at 300 K were performed after every energy minimization. In each molecular dynamics simulation, the dynamic step was 0.001 ps and total simulation time was 700 steps (0.7 ps). Each peptide was run as a neutral or appropriately charged molecule on both a flat (001) apatite plane and a [010] step introduced on the (001) plane (fig. 1). Four different initial start orientations were simulated for each peptide on both the flat and step lattices.
Fig. 1.
Flat (001) and hexagonal [010] step in (001) lattices used in calculating peptide adsorption energies.
Results
Nineteen phage sequences emerged multiple times after DNA sequencing. Of the 19 clones, 17 were found in more than 1 experiment on the same substrate, either BLM or HA. Of these 17 phage clones, 7 had high affinity towards both BLM and HA (table 1). In all phage display experiments, the streptavidin control yielded the HPQ consensus sequence signifying successful panning.
Table 1.
Phage sequences appearing on both BLM and HA
| Phage sequences | Total clones sequenced, % |
|---|---|
| APWHLSSQYSRT* | 21.8 |
| IDTFYMSTMSHS*, ^ | 8.6 |
| ALTLHPQPLDHP* | 4.9 |
| TALATSSTYDPH | 4.9 |
| VTKHLNQISQSY*,^ | 2.5 |
| WSSGMTPDTGAP^ | 2.5 |
| ALSSSSNTTTRV | 1.6 |
Clone ran in modified ELISA.
Identified as a high-information clone in INFO.
All phage sequences were analyzed with the INFO program provided by RELIC. Compared to the NEB background phage set, the phage set identified as adsorbing to the HA and/or BLM showed a trend towards higher information content. Regions of greater occurrence were partitioned, and the partitioned phage lists were cross-referenced with repeat phage lists. Of the 19 phage sequences appearing multiple times, over 60% were located in a partitioned region of high information (table 1).
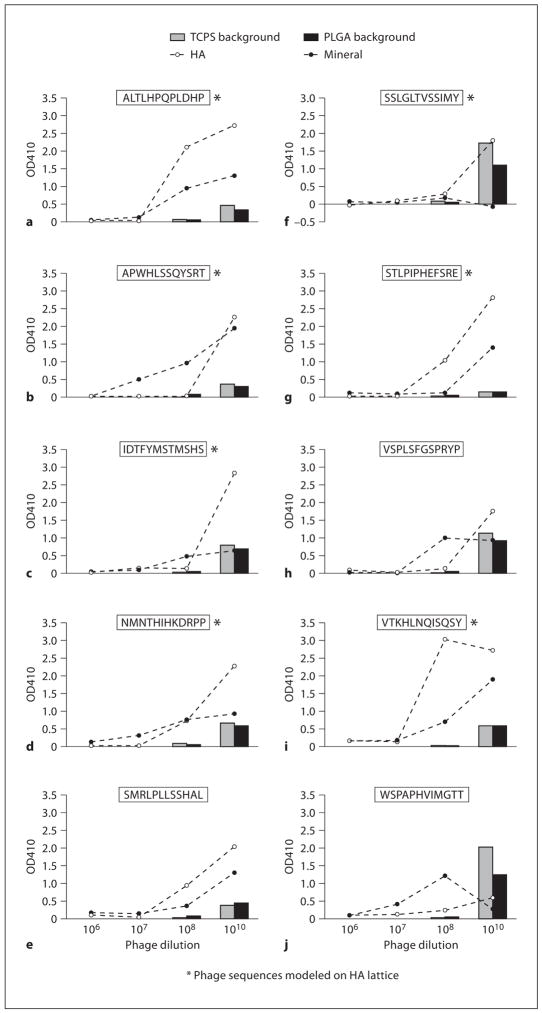
Of the 10 phage clones chosen for qualitative ELISA, 7 occurred as repeats, whereas 3 (STLPIPHEFSRE, VSPLSFGSPRYP and WSPAPHVIMGTT) occurred only once. Phage dilutions of 108 and 1010 plaque-forming units resulted in positive ELISA readings, with a stronger positive signal for the 1010 dilution (fig. 2). TCPS and PLGA films showed consistent background adsorption for all peptides and concentrations. Low adsorption of the selected phage to TCPS and PLGA compared to HA and BLM illustrates the identified phage have specificity to apatite-based substrates. Favorable phage binding was observed for all sequences except SSLGLTVSSIMY, VSPLSFGSPRYP and WSPAPHVIMGTT (fig. 2f, h, j).
Fig. 2.
Results of modified ELISA of 10 phage clones on BLM, HA, TCPS and PLGA. High adsorption to BLM and HA in addition to low adsorption to TCPS and PLGA supports that the clones isolated on apatite-based materials are substrate specific.
For neutral peptides, rank of peptide adsorption energy was consistent between the (001) plane and the [010] step (table 2). For charged peptides, a less consistent trend in peptide rank was observed. Rather, for the charged peptides on the [010] step, greater adsorption energies were calculated for peptides with low isoelectric points. To decide which peptides to synthesize for further investigation, the peptides ranking within the top 5 in both the neutral and charged peptide scenarios were evaluated along with the RELIC analysis, modified ELISA results and clone frequency results over the 3 phage experiments. The peptide sequences with the greatest overall potential for adsorption on HA disks and BLM films include APWHLSSQYSRT, STLPIPHEFSRE and VTKHLNQISQSY.
Table 2.
Peptide adsorption energies for 19 repeat peptide sequences calculated from molecular model simulations on the (001) HA face and the [010] step on (001) HA face for both charged and neutral peptides
| Adsorption on (001) plane |
Adsorption on [010] step on (001) plane |
||||||
|---|---|---|---|---|---|---|---|
| rank | peptide | peptide adsorption energy, kcal/mol | pI | rank | peptide | peptide adsorption energy, kcal/mol | pI |
| Neutral peptides | |||||||
| 1 | vtkhlnqisqsy1 | −336.9 | 9.7 | 1 | gtlseklrdtha | −727.9 | 7.8 |
| 2 | vnysmeiplvps | −304.9 | 3.3 | 2 | vnysmeiplvps | −713.9 | 3.3 |
| 3 | alssssntttrv | −288.1 | 11.1 | 3 | vtkhlnqisqsy1 | −705.6 | 9.7 |
| 4 | idtfymstmshs1 | −284.4 | 4.9 | 4 | idtfymstmshs1 | −696.5 | 4.9 |
| 5 | apwhlssqysrt1 | −279.6 | 9.9 | 5 | talatsstydph | −688.0 | 4.9 |
| 6 | gtlseklrdtha | −275.7 | 7.8 | 6 | alssssntttrv | −684.0 | 11.1 |
| 7 | talatsstydph | −266.8 | 4.9 | 7 | altlhpqpldhp1 | −683.9 | 6.0 |
| 8 | hgevprfhavhl | −260.7 | 8.1 | 8 | apwhlssqysrt1 | −680.8 | 9.9 |
| 9 | nmnthihkdrpp1 | −249.5 | 10.1 | 9 | stlpiphefsre1 | −677.3 | 5.3 |
| 10 | gapalstpplsr | −247.5 | 11.1 | 10 | hnffpetpssgp | −657.3 | 5.1 |
| 11 | stlpiphefsre1 | −239.3 | 5.3 | 11 | hhnvhlltsgnq | −654.8 | 8.1 |
| 12 | sslgltvssimy1 | −233.9 | 5.9 | 12 | hgevprfhavhl | −653.6 | 8.1 |
| 13 | hnpkamfygvns | −231.0 | 9.7 | 13 | wssgmtpdtgap | −622.1 | 3.1 |
| 14 | yqtsspakqsvg | −227.5 | 9.7 | 14 | qnmmspiegvri | −610.7 | 7.0 |
| 15 | qnmmspiegvri | −223.2 | 7.0 | 15 | nmnthihkdrpp1 | −598.2 | 10.1 |
| 16 | hhnvhlltsgnq | −209.0 | 8.1 | 16 | sslgltvssimy1 | −590.8 | 5.9 |
| 17 | hnffpetpssgp | −197.6 | 5.1 | 17 | gapalstpplsr | −582.7 | 11.1 |
| 18 | wssgmtpdtgap | −190.6 | 3.1 | 18 | yqtsspakqsvg | −569.0 | 9.7 |
| 19 | altlhpqpldhp1 | −187.1 | 6.0 | 19 | hnpkamfygvns | −564.7 | 9.7 |
|
| |||||||
| Charged peptides | |||||||
| 1 | alssssntttrv | −388.3 | 11.1 | 1 | idtfymstmshs1 | −872.0 | 4.9 |
| 2 | stlpiphefsre1 | −269.4 | 5.3 | 2 | talatsstydph | −848.7 | 4.9 |
| 3 | idtfymstmshs1 | −263.1 | 4.9 | 3 | wssgmtpdtgap | −848.3 | 3.1 |
| 4 | talatsstydph | −260.7 | 4.9 | 4 | hnffpetpssgp | −822.7 | 5.1 |
| 5 | altlhpqpldhp1 | −251.3 | 6.0 | 5 | stlpiphefsre1 | −794.8 | 5.3 |
| 6 | apwhlssqysrt1 | −248.8 | 9.9 | 6 | altlhpqpldhp1 | −713.4 | 6.0 |
| 7 | hnffpetpssgp | −239.7 | 5.1 | 7 | hhnvhlltsgnq | −704.5 | 8.1 |
| 8 | wssgmtpdtgap | −212.3 | 3.1 | 8 | hgevprfhavhl | −629.2 | 8.1 |
| 9 | qnmmspiegvri | −205.2 | 7.0 | 9 | qnmmspiegvri | −620.0 | 7.0 |
| 10 | hhnvhlltsgnq | −190.5 | 8.1 | 10 | sslgltvssimy1 | −586.2 | 5.9 |
| 11 | gapalstpplsr | −179.5 | 11.1 | 11 | apwhlssqysrt1 | −545.6 | 9.9 |
| 12 | sslgltvssimy1 | −178.8 | 5.9 | 12 | vnysmeiplvps | −507.9 | 3.3 |
| 13 | gtlseklrdtha | −173.0 | 7.8 | 13 | vtkhlnqisqsy1 | −495.2 | 9.7 |
| 14 | nmnthihkdrpp1 | −160.6 | 10.1 | 14 | gtlseklrdtha | −471.2 | 7.8 |
| 15 | vtkhlnqisqsy1 | −145.2 | 9.7 | 15 | nmnthihkdrpp1 | −470.2 | 10.1 |
| 16 | hgevprfhavhl | −136.8 | 8.1 | 16 | hnpkamfygvns | −421.0 | 9.7 |
| 17 | hnpkamfygvns | −114.7 | 9.7 | 17 | yqtsspakqsvg | −419.7 | 9.7 |
| 18 | yqtsspakqsvg | −113.4 | 9.7 | 18 | alssssntttrv | −405.8 | 11.1 |
| 19 | vnysmeiplvps | 0.0 | 3.3 | 19 | gapalstpplsr | 0.0 | 11.1 |
pI = Isoelectric points.
Clone ran in ELISA.
Discussion
Phage display libraries, presenting approximately 109 different combinations of linear peptide inserts, have isolated high-affinity sequences with specificity towards substrate materials from semiconductors [Whaley et al., 2000] and plastics [Adey et al., 1995] to microglial [Samoylova et al., 2002] and neuroblastoma tumor cells [Zhang et al., 2001]. To our knowledge, this work is the first to report peptide sequences identified via phage display on BLM. A novel multidisciplinary approach of combining benchtop phage display results with observations of peptide-apatite interactions through molecular modeling has been introduced.
Several methods of analysis were performed to validate the interactions of identified peptides on apatite-based materials. The INFO program used reflects the possibility that each individual peptide sequence is identified by chance, by calculating an information measure as the negative natural log of each peptide’s probability of being found in the native library. A low-information measure can be indicative of phage sequences likely to appear in the population based on amino acid composition and/or with high growth rates during amplification. A high-information clone can indicate an uncommon sequence with a low growth rate. The sequence STLPIPHEFSRE occurred once in the 3 phage display experiments; however, the peptide was identified as a high-information clone and exhibited positive binding in the modified ELISA.
The modified ELISA tested for the specific interaction of each of the identified phage with the 4 materials examined and produced evidence that the majority of phages isolated from panning were specific towards the apatite-based substrates. Unfavorable binding resulted when the background signal was greater than or equal to the phage adsorption signal (fig. 2f, h, j). Even though phage with a high background signal could still show specificity towards BLM and HA, these sequences were not considered for future in vitro experiments.
Modeling organic molecules has proven useful in a variety of organic-inorganic systems including the investigation of protein-substrate and peptide-substrate interactions in calcite and calcium oxalate [Qiu et al., 2004]. Despite a basis of modeling work on the apatite lattice alone [Mkhonto and de Leeuw, 2002; Hauptmann et al., 2003; Astala and Stott, 2005], limited studies have modeled organic molecules on an apatite lattice [Fujisawa, 1998]. Using Newtonian physics in a time-dependent manner to calculate forces on a system, molecular mechanics simulations allow the calculation of peptide adsorption energies. A greater negative peptide adsorption energy translates into a peptide possessing more potential to adsorb to the lattice. When a step is introduced to the hydroxyapatite lattice, adsorption energies increase, likely because of the altered charge and higher degree of undercoordination of atoms and molecules on the surface of the step. The effect of negatively charged amino acid sequences on adsorption has been suggested as one mechanism of protein/peptide adsorption to hydroxyapatite [Fujisawa et al., 1997]. These data provide molecular modeling evidence that, given appropriate charge for a neutral pH environment, acidic peptides exhibit favorable binding over basic peptides. Furthermore, the model system used in these experiments provides a high-throughput validation technique where multiple interactions can be investigated prior to further in vitro or in vivo testing.
The phage panning on apatite-based materials yielded both acidic and basic peptide sequences, which is counterintuitive to the idea that apatite attracts mainly acidic sequences. The peptide sequences from phage chosen as having the greatest overall potential for adsorption on BLM and HA include APWHLSSQYSRT, STLPIPHEFSRE and VTKHLNQISQSY. The peptide VTKHLNQISQSY is similar in composition to regions found in fibromodulin, lumican and decorin.
Development of bioactive organic-mineral hybrid materials that promote bone regeneration continues to advance. Ongoing efforts aim to identify peptide sequences via phage display with high affinity towards a specific adult stem cell population. Employing the dual peptide method to anchor the peptide on apatite-like material while binding to cells capable of bone differentiation will be a powerful biomolecular approach to improve bone regeneration.
Acknowledgments
This work was supported by NIH R01 DE 013380 and DE 015411 (D.H.K.), Tissue Engineering at Michigan Grant DE 07057 (S.S.) and the NIRT program of the NSF (EAR-0403732) (S.B., U.B.). The authors would like to thank Constance Esposito for help with DNA sequencing and Alisha Diggs for help with HA disk preparation.
Abbreviations used in this paper
- BLM
bone-like mineral
- ELISA
enzyme-linked immunosorbent assay
- HA
hydroxyapatite
- mSBF
modified simulated body fluid
- NEB
New England Biolabs
- PLGA
polylactic-co-glycolic acid
- TCPS
tissue culture polystyrene
References
- Adey NB, Mataragnon AH, Rider JE, Carter JM, Kay BK. Characterization of phage that bind plastic from phage-displayed random peptide libraries. Gene. 1995;156:27–31. doi: 10.1016/0378-1119(95)00058-e. [DOI] [PubMed] [Google Scholar]
- Alsberg E, Hill EE, Mooney DJ. Craniofacial tissue engineering. Crit Rev Oral Biol Med. 2001;12:64–75. doi: 10.1177/10454411010120010501. [DOI] [PubMed] [Google Scholar]
- Astala R, Stott MJ. First principles investigation of mineral component of bone: CO3 substitutions in hydroxyapatite. Chem Mat. 2005;17:4125–4133. [Google Scholar]
- Damsky CH. Extracellular matrixintegrin interactions in osteoblast function and tissue remodeling. Bone. 1999;25:95–96. doi: 10.1016/s8756-3282(99)00106-4. [DOI] [PubMed] [Google Scholar]
- Feinberg SE, Aghaloo TL, Cunningham LL., Jr Role of tissue engineering in oral and maxillofacial reconstruction: findings of the 2005 AAOMS research summit. J Oral Maxillofac Surg. 2005;63:1418–1425. doi: 10.1016/j.joms.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Fujisawa R, Mizuno M, Nodasaka Y, Kuboki Y. Attachment of osteoblastic cells to hydroxyapatite crystals by a synthetic peptide (Glu7-Pro-Arg-Gly-Asp-Thr) containing two functional sequences of bone sialoprotein. Matrix Biol. 1997;16:21–28. doi: 10.1016/s0945-053x(97)90113-x. [DOI] [PubMed] [Google Scholar]
- Fujisawa R, Kuboki Y. Conformation of dentin phosphophoryn adsorbed on hydroxyapatite crystals. Eur J Oral Sci. 1998;106(suppl 1):249–253. doi: 10.1111/j.1600-0722.1998.tb02183.x. [DOI] [PubMed] [Google Scholar]
- Garcia AJ, Reyes CD. Bio-adhesive surfaces to promote osteoblast differentiation and bone formation. J Dent Res. 2005;84:407–413. doi: 10.1177/154405910508400502. [DOI] [PubMed] [Google Scholar]
- Hauptmann S, Dufner H, Brickmann J, Kast SM, Berry RS. Potential energy function for apatites. Phys Chem Chem Phys. 2003;5:635–639. [Google Scholar]
- Luong LN, Hong SI, Patel RJ, Outslay ME, Kohn DH. Spatial control of protein within biomimetically nucleated mineral. Biomaterials. 2006;27:1175–1186. doi: 10.1016/j.biomaterials.2005.07.043. [DOI] [PubMed] [Google Scholar]
- Mandava S, Makowski L, Devarapalli S, Uzubell J, Rodi DJ. RELIC – a bioinformatics server for combinatorial peptide analysis and identification of protein-ligand interaction sites. Proteomics. 2004;4:1439–1460. doi: 10.1002/pmic.200300680. [DOI] [PubMed] [Google Scholar]
- Mkhonto D, de Leeuw NH. A computer modelling study of the effect of water on the surface structure and morphology of fluorapatite: introducing a Ca10(PO4)6F2 potential model. J Mater Chem. 2002;12:2633–2642. [Google Scholar]
- Muschler GF, Midura RJ. Connective tissue progenitors: practical concepts for clinical applications. Clin Orthop Relat Res. 2002;395:66–80. doi: 10.1097/00003086-200202000-00008. [DOI] [PubMed] [Google Scholar]
- Qiu SR, Wierzbicki A, Orme CA, Cody AM, Hoyer JR, Nancollas GH, Zepeda S, De Yoreo JJ. Molecular modulation of calcium oxalate crystallization by osteopontin and citrate. Proc Natl Acad Sci USA. 2004;101:1811–1815. doi: 10.1073/pnas.0307900100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappe AK, Goddard WA., III Charge equilibrium for molecular dynamics simulations. J Phys Chem. 1991;95:3358–3363. [Google Scholar]
- Rappe AK, Casewit CJ, Colwell KS, Goddard WA, Skiff WM. UFF, a full periodic-table force field for molecular-mechanics and molecular-dynamics simulations. J Am Chem Soc. 1992;114:10024–10035. [Google Scholar]
- Samoylova TI, Ahmed BY, Vodyanoy V, Morrison NE, Samoylov AM, Globa LP, Baker HJ, Cox NR. Targeting peptides for microglia identified via phage display. J Neuroimmunol. 2002;127:13–21. doi: 10.1016/s0165-5728(02)00071-1. [DOI] [PubMed] [Google Scholar]
- Segvich S, Smith HC, Luong LN, Kohn DH. Uniform deposition of protein incorporated mineral layer on three-dimensional porous polymer scaffolds. J Biomed Mater Res B Appl Biomater. 2008;84B:340–349. doi: 10.1002/jbm.b.30877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whaley SR, English DS, Hu EL, Barbara PF, Belcher AM. Selection of peptides with semiconductor binding specificity for directed nanocrystal assembly. Nature. 2000;405:665–668. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]
- Wiesmann HP, Joos U, Meyer U. Biological and biophysical principles in extra-corporeal bone tissue engineering. Part II. Int J Oral Maxillofac Surg. 2004;33:523–530. doi: 10.1016/j.ijom.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Yaszemski MJ, Payne RG, Hayes WC, Langer R, Mikos AG. Evolution of bone transplantation: molecular, cellular and tissue strategies to engineer human bone. Biomaterials. 1996;17:175–185. doi: 10.1016/0142-9612(96)85762-0. [DOI] [PubMed] [Google Scholar]
- Zhang J, Spring H, Schwab M. Neuroblastoma tumor cell-binding peptides identified through random peptide phage display. Cancer Lett. 2001;171:153–164. doi: 10.1016/s0304-3835(01)00575-4. [DOI] [PubMed] [Google Scholar]