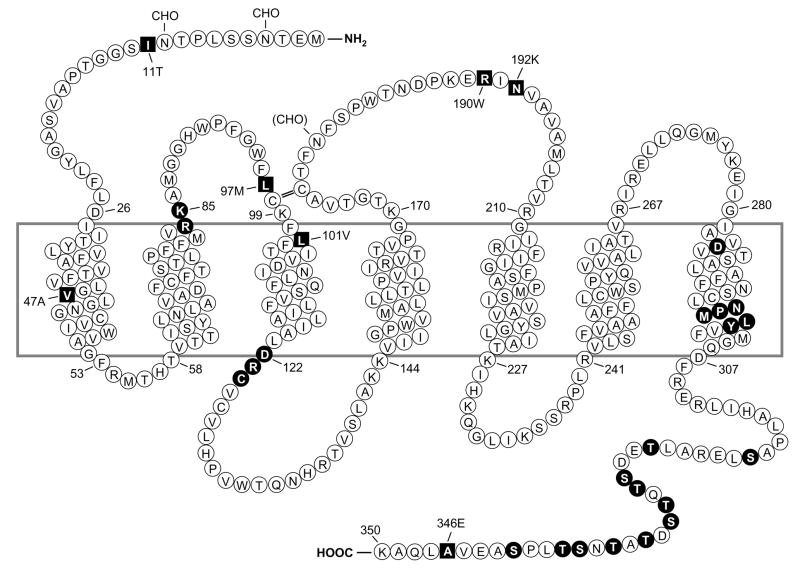
Figure 1. Predicted transmembrane disposition of the human FPR1.
The protein sequence of the FPR-98 isoform (L110, A346) is shown (Boulay, 1990a). The transmembrane domains (TMs) are predicted based on hydrophobicity of the amino acid sequence and on similarities to the rhodopsin structure. The amino acids that form the boundaries of the TMs are numbered. One-letter amino acid code is used. The square blocks in revserce color represent positions where amino acid substitutions result from polymorphisms, including amino acids 11 (I/T), 47 (V/A), 101 (L/V), 190 (R/W), 192 (N/K) and 346 (A/E). The circle blocks in reverse color indicate amino acids with known functions as follows. R84, K85 and D284 are critical for high-affinity binding of fMLF (Mills et al., 1998; Quehenberger et al., 1997). D122, R123 and C124 are the signature sequence for G protein interaction (DRY in many GPCRs). NPMLY in the TM VII are known signature sequence (NPXXY) for receptor internalization (Gripentrog et al., 2000; He et al., 2001). The 11 Ser and Thr residues in the cytoplasmic tail are potential phosphorylation sites for G protein-coupled receptor kinase 2 and 3 (GRK2, GRK3) (Prossnitz et al., 1995). CHO, carbohydrate, marks the identified and potential (in parenthesis) sites for N-glycosylation. The predicted disulfide bond between C98 and C176 is marked with double-line (=).