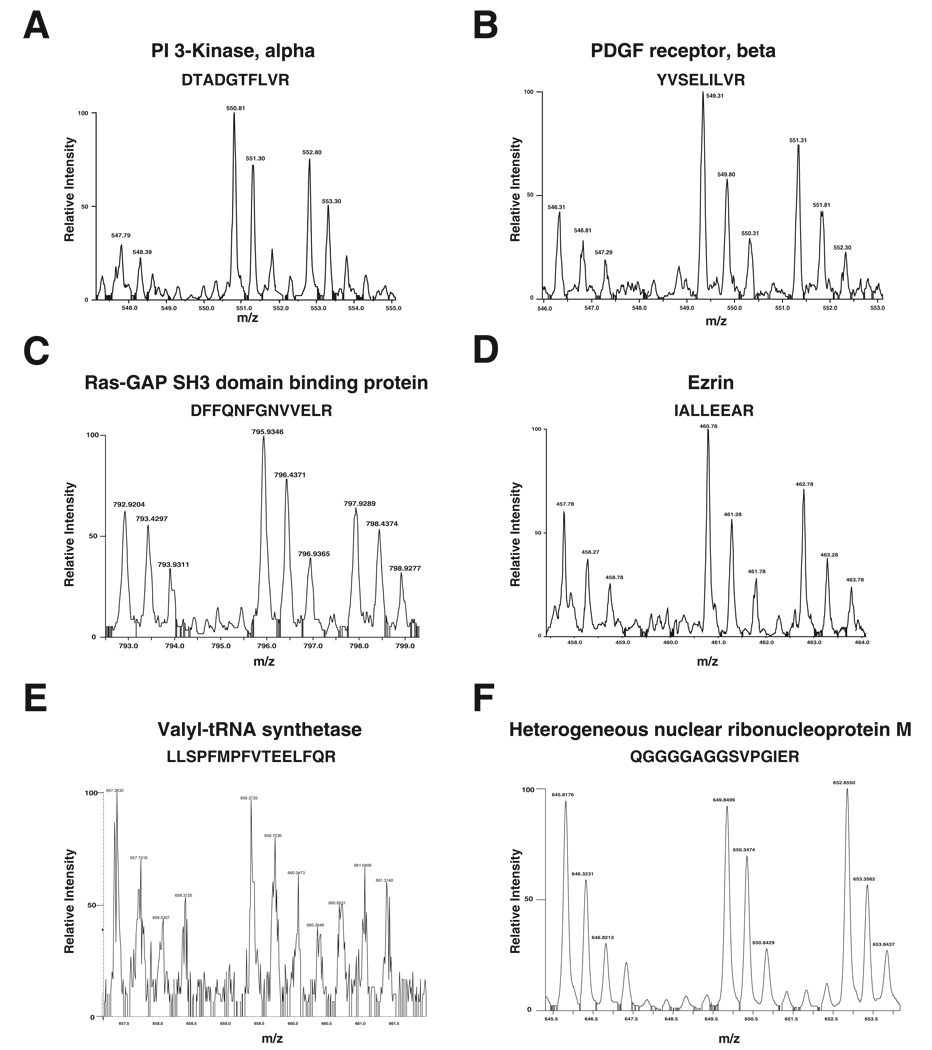
Figure 3.
MS spectra of proteins showing different profiles identified in our SILAC screen. The 3 spectral peaks in each figure represent the mass shift of the same peptide. (A) A doubly charged peptide with the sequence DTADGTFLVR, from PI 3-Kinase, alpha (1: 3.3: 2.2); (B) A doubly charged peptide with the sequence YVSELILVR from PDGF receptor, beta (1: 2: 1.5); (C) A doubly charged peptide with the sequence DFFQNFGNVVELR from Ras-GAP SH3 domain containing protein (1: 1.5: 1); (D) A doubly charged peptide with the sequence IALLEEAR from Ezrin (1: 1.6: 1.2); (E) A triply charged peptide with the sequence LLSPFMPFVTEELFQR from valyl-tRNA synthetase (1: 1.03: 0.74); (F) A doubly charged peptide with the sequence from QGGGGAGGSVPGIER from heterogeneous nuclear ribonucleoprotein M (1: 1.1: 1.1). While some of the proteins (A–D) show involvement in PDGF signaling pathway as c-Src substrates, some proteins do not show any involvement in PDGF signaling pathway (E) or do not show characteristic spectra as that of c-Src substrates (F). The ratios shown in parentheses reflect ratio of abundance of peptides across three states (light: medium: heavy).