Fig. 6.
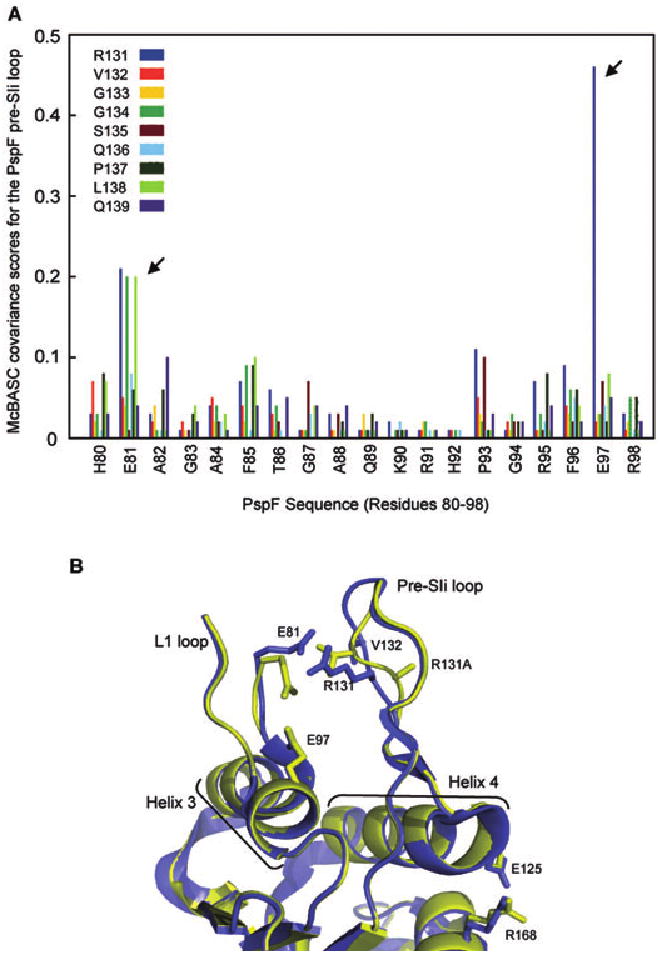
A conserved switch between the pre-SIi and L1 loops exists within bEBPs.
A. The covariance between the PspF pre-SIi sequence (RVGGSQPLQ; colour-coded as shown) and PspF residues 80–98 was calculated and depicted graphically. The strong covariance of residue E81 with pre-SIi residues R131, G134 and L138 (arrowed) and residue E97 and pre-SIi residue R131 (arrowed) is indicated by high covariance scores.
B. The crystal structures of the apo-PspF1–275R131A (yellow) and apo-PspF1–275WT (blue; PDB 2BJW) demonstrate the effect of the R131A mutation on the pre-SIi loop conformation. The two structures were aligned on the main chain atoms of residues 35–42. The positions of residues E81 (L1 loop), E125 (Helix 4), R168 (putative R-finger), R131 and V132 (pre-SIi) and the R131A mutation (in the context of the PspF1–275R131A structure) are indicated. Structural features relevant to bEBPs such as the L1 and pre-SIi loops, Helix 3 and Helix 4 are labelled. Clear local differences between the apo-PspF1–275WT and apo-PspF1–275R131A structures are apparent in the pre-SIi loop conformation, as well as a significant rotation of residue E81 (L1 loop) resulting in the disruption of the E81-R131 link.
