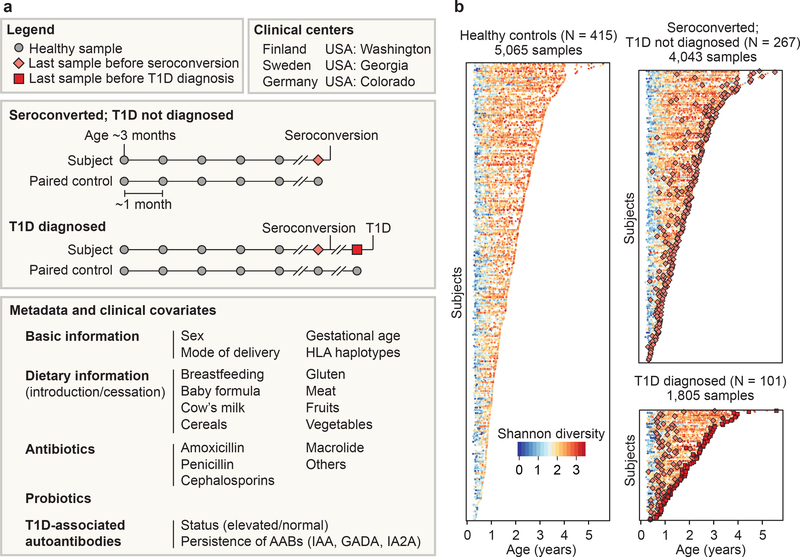
Figure 1: >10,000 longitudinal gut metagenomes from the TEDDY T1D cohort.
We analyzed 10,913 metagenomes collected longitudinally from 783 children (415 controls, 267 seroconverters, and 101 diagnosed with T1D) approximately monthly over the first five years of life. a, Subjects were recruited at six clinical centers. Primary endpoints were seroconversion (defined as persistent confirmed IA) and T1D diagnosis. Additional metadata analyzed for subjects and samples included breastfeeding status, birth mode, probiotics, antibiotics, formula feeding, and other dietary covariates. b, Overview of stool samples collected and microbiome development as summarized by Shannon alpha-diversity and stratified by endpoint. Median number of samples per individual N = 12 (healthy controls N = 10, seroconverters N = 13, T1D cases N = 16).