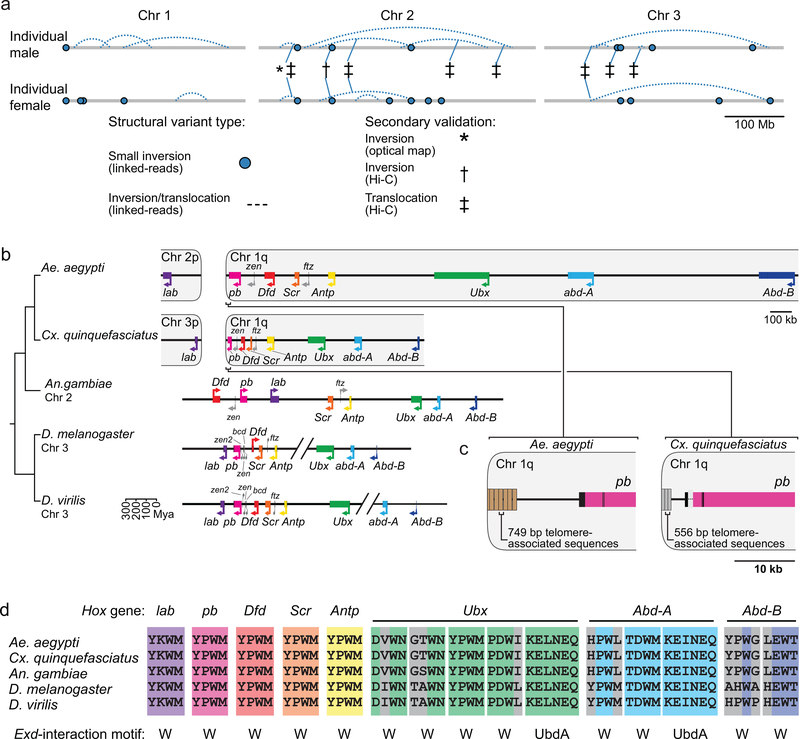
Extended Data Figure 8 |. Structural variation and Hox cofactor motifs.
a, linked-read sequencing of two individuals from the LVP_AGWG strain identified putative structural variants in the AaegL5 assembly. b, Comparative genomic arrangement of the Hox cluster (HOXC) in 5 species (Supplementary Data 22). Due to chromosome arm exchange, Chr 3p in Cx. quinquefasciatus is the homologue of Chr 2p in Ae. aegypti6. c, Repeats in putative telomere-associated sequences downstream of pb in both species. d, Motifs known to mediate protein-protein interactions with the Hox cofactor Extradenticle (Exd)91 from the five indicated species are aligned using Clustal-Omega. Perfectly aligned residues are coloured according to Hox gene identity, non-conserved residues are grey.